You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Li-Yen Chang.
Nipah virus (NiV) is a highly lethal zoonotic paramyxovirus that emerged in Malaysia in 1998. It is a human pathogen capable of causing severe respiratory infection and encephalitis. The natural reservoir of NiV, Pteropus fruit bats, remains a continuous virus source for future outbreaks, although infection in the bats is largely asymptomatic. NiV provokes serious disease in various mammalian species.
- henipavirus infections
- encephalitis
- chiroptera
1. Introduction
In the events of the recent COVID-19 pandemic, more attention and effort has been devoted to studies and research on potential pandemic-causing pathogens, one of which is the Nipah virus (NiV). NiV is an emerging paramyxovirus with a high pathogenicity that has been causing near-annual outbreaks in the South Asia region since its discovery in Malaysia in 1998 [1]. It is currently listed as one of the top 10 emerging viruses that require urgent research and development in public health emergency contexts by the World Health Organization (WHO) [2], and it has been made a priority for vaccine development by the Coalition for Epidemic Preparedness Innovations (CEPI) [3] and the United Kingdom Vaccine Network [4].
NiV is a negative-sense single-stranded RNA enveloped virus and is a member of the genus Henipavirus in the family Paramyxoviridae [5,6][5][6]. The genome of the virus is non-segmented and is approximately 18 kb nucleotides long [6,7,8,9][6][7][8][9]. The viral genome encodes six structural proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), fusion glycoprotein (F), attachment glycoprotein (G) and the RNA polymerase or large protein (L). In addition, there are three accessory proteins within the P: the V, W and C proteins, as a result of mRNA editing and the alternative start codon. Overall, NiV genome sequence analyses have identified two main clades: the M genotype, which comprises the Malaysian NiV isolates (NiV-M), and the B genotype, which includes Bangladesh (NiV-B) and India NiV isolates (NiV-I) [7,10,11][7][10][11]. Despite the three strains sharing a high percentage of homology (NiV-M and NiV-B strains share 91.8% homology, and NiV-I sharing 85.14–96.15% homology with both NiV-M and NiV-B), the B clade infections were shown to be significantly more pathogenic than the M clade [10,12,13,14][10][12][13][14].
In the Malaysia outbreak, NiV infection was characterized as a respiratory and neurological disease that resulted in over 250 cases and fatalities exceeding 100 cases [1]. Besides Malaysia, NiV was reported in neighboring countries such as Singapore, the Philippines and South Asia (Bangladesh and India). In South Asia, cases were reported almost annually, with the most recent case of NiV infection reported in 2021 in Kerala, India [15]. Clinically, respiratory infections were more common and the mortality was higher among the NiV cases reported in Bangladesh and India, as compared to the cases in South East Asia. The variation in severity of symptoms and mortality between NiV cases in both regions could be attributed to the different genetic makeup of the strains pervasive in either region, or the differences in access and quality of medical care between the two regions [16]. Nevertheless, NiV infections are generally associated with acute respiratory distress, encephalitis and in some cases myocarditis [17]. Additionally, some patients experienced drowsiness, extreme lethargy, mental confusion and in the worst cases coma. In fact, a percentage of patients experienced residual neurological complications, such as late-onset encephalitis, years after the initial infection [18,19][18][19].
Fruit bats of the Pteropus genus have been suggested as the natural reservoir for NiV. The bats harboring NiV remain asymptomatic [20,21,22][20][21][22] and therefore could facilitate the spread of the virus to susceptible hosts during spillover events. In humans, epidemiological studies have implicated animal-to-human and human-to-human transmissions as the main routes of NiV spread; the former was connected to exposure to infected animal fluids such as its saliva, urine and excreta, whereas the latter was connected through contact with body fluids from infected individuals, specifically via respiratory droplets [20,23,24][20][23][24]. The route of NiV transmission in the Malaysia and Singapore outbreaks was identified to be animal-to-human, whereby bats harboring NiV transmitted the virus to pigs through direct contact, which then acted as amplifying hosts and subsequently transmitted the virus to humans via aerosol droplets [1,5][1][5]. Meanwhile, in the Bangladesh and India NiV outbreaks that occur almost annually, transmission of the virus is also animal-to-human, but via ingestion of food or fluid contaminated by NiV-infected bats or via direct contact with NiV-infected bats. Besides this, the human-to-human transmission of NiV was also reported, and this was a common mode of transmission in Bangladesh, comprising half of the NiV cases reported between 2001 and 2007 [25,26,27][25][26][27]. Sociocultural expectations to care for ill family members, poor infection control practices and lack of healthcare resources are factors that could have contributed to the higher number of human-to-human NiV transmissions in Bangladesh relative to Malaysia [24,26][24][26].
Despite NiV outbreaks occurring almost annually and the pandemic potentiality of the virus, no vaccines or therapeutics have yet been approved and made available for human use [28,29,30][28][29][30]. Vaccines in general are targeted to induce humoral immunity, specifically protective antibodies; recent vaccine development also aims to generate cellular immunity. This is because both immune subsystems are crucial to provide an effective immune response towards the infection and for protection against the disease. However, inadequate clinical specimens available for in-depth analysis due to no following NiV outbreak in Malaysia and small sporadic outbreaks of NiV in Bangladesh and India result in constraints to the recapitulation of clinical signs of the human NiV disease, as well as the monitoring and evaluation of the immune response following NiV exposure.
2. Replication Cycle of NiV
The NiV particle has six structural proteins, namely the N, P, M, F, G and L, which are arranged accordingly in the RNA genome from 3′ to 5′ (Figure 1) [6]. The replication cycle of NiV starts when the virion attaches to the host cell receptors, ephrin-B2 and -B3 via the NiV G protein [31,32,33][31][32][33]. Next, the NiV F protein mediates the fusion of the viral envelope with the host cell membrane, releasing the viral genome into the cytoplasm. The viral genomic RNA is associated with N, P and L proteins, which forms the ribonucleoprotein complex and is involved in the transcription and replication of the virus. The L polymerase catalyzes the transcription of the virus genomic RNA into mRNAs for protein translation. The translated viral surface glycoproteins F and G are inserted into the host cell endoplasmic reticulum for post-translational modifications, particularly glycosylation. The other translated viral proteins—N, P, M and L—remain in the cytoplasm. When abundant viral mRNA transcripts are produced, full-length anti-genomes are then synthesized to generate more copies of the NiV genome. These new copies of genome assemble with the viral proteins near the host cell membrane where F and G proteins are studded, and the budding of new virions facilitated by the M protein will occur.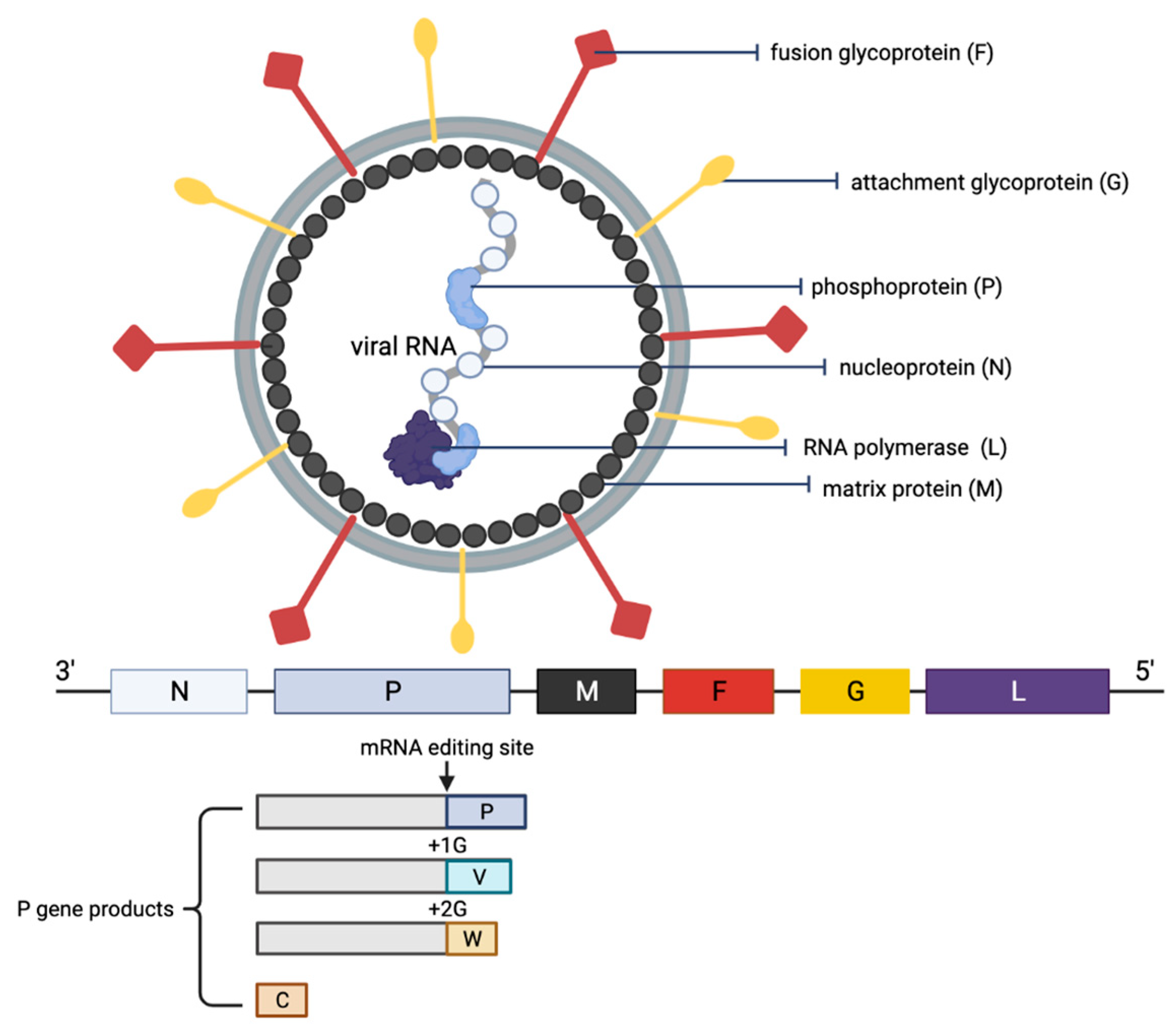
Figure 1. Schematic representation of the structure of an NiV particle and the viral genome organization. The NiV N, P and L proteins interact with the viral RNA to form the ribonucleoprotein complex, which is surrounded by a lipid bilayer envelope containing the NiV glycoproteins F and G. The NiV M protein is associated with the inner side of the envelope. The viral proteins and arrangement of genes in the viral genome from 3′–5′ are color-coded, respectively, for identification. The NiV P gene products (V, W and C proteins) as a result of mRNA editing are illustrated. The V protein contains a single G insertion, and translation shifts it to +1 reading frame. The W protein contains two G insertions, shifting the translation to the +2 reading frame. The C protein is translated from an internal open reading frame of the P gene.
3. Pathogenesis of NV
NiV enters through the oronasal route into human and other animal hosts to cause an infection. The virus infects the epithelium cells along the respiratory tract, and a high concentration of viral antigens could be detected in the lymphoid and respiratory tissues [12]. Initial viremia then spreads the virus to other parts of the body, while secondary replication occurs in the endothelium. The NiV infection of host cells starts when the viral G protein attaches to the cellular receptors ephrin-B2 and -B3 [31,32,33][31][32][33]. The virus then rapidly disseminates to different organs, including the spleen, kidneys, heart and liver within the first week of infection [14,34,35][14][34][35]. Both ephrin-B2 and -B3 are found on a wide range of cell types including epithelial and endothelial cells, as well as neurons. Both these cellular receptors are highly conserved across animal species, which explains the broad species and tissue tropism of NiV [36]. Interestingly, a recent study has observed that smooth muscle cells that lack the cellular receptors ephrin-B2 and -B3 were permissive to NiV infection and produced high viral titers similar to permissive cells expressing the cellular receptors [37]. There was prolonged NiV production in the smooth muscle cells with no cytopathogenic effects. Together, the study suggested the likely existence of an unidentified entry receptor for NiV or a non-specific virus entry mechanism. Besides, NiV was also reported to enter and infect the central nervous system via circulating immune cells, specifically immature dendritic cells and monocytic cells [38]. These cells were noted to be NiV-permissive; however, the virus did not replicate efficiently in them. Nevertheless, the NiV-infected immune cells migrated across the in vitro blood–brain barrier and infected susceptible cells in a focused manner, similar to observed neuronal infection and the presence of focal lesions in the brain of both NiV-infected human and animals [39,40][39][40].References
- Chua, K.B.; Goh, K.J.; Wong, K.T.; Kamarulzaman, A.; Seow, P.; Tan, K.; Ksiazek, T.G.; Zaki, S.R.; Paul, G.; Lam, S.K.; et al. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 1999, 354, 1257–1259.
- World Health Organization. WHO R&D Nipah Baseline Situation Analysis; WHO: Geneva, Switzerland, 2018; p. 41.
- CEPI. Priority Diseases. Available online: https://cepi.net/research_dev/priority-diseases/ (accessed on 31 March 2022).
- UK Vaccine Network. Available online: https://www.gov.uk/government/groups/uk-vaccines-network (accessed on 31 March 2022).
- Chua, K.; Bellini, W.; Rota, P.; Harcourt, B.; Tamin, A.; Lam, S.K.; Ksiazek, T.G.; Rollin, P.E.; Zaki, S.R.; Shieh, W.J.; et al. Nipah virus: A recently emergent deadly paramyxovirus. Science 2000, 288, 1432–1436.
- Harcourt, B.H.; Tamin, A.; Ksiazek, T.G.; Rollin, P.E.; Anderson, L.J.; Bellini, W.J.; Rota, P.A. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 2000, 271, 334–349.
- Harcourt, B.H.; Lowe, L.; Tamin, A.; Liu, X.; Bankamp, B.; Bowden, N.; Rollin, P.E.; Comer, J.A.; Ksiazek, T.G.; Hossain, M.J.; et al. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg. Infect. Dis. 2005, 11, 1594–1597.
- Chan, Y.P.; Chua, K.B.; Koh, C.L.; Lim, M.E.; Lam, S.K. Complete nucleotide sequences of Nipah virus isolates from Malaysia. J. Gen. Virol. 2001, 82, 2151–2155.
- Wang, L.F.; Harcourt, B.H.; Yu, M.; Tamin, A.; Rota, P.A.; Bellini, W.J.; Eaton, B.T. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 2001, 3, 279–287.
- Yadav, P.D.; Shete, A.M.; Kumar, G.A.; Sarkale, P.; Sahay, R.R.; Radhakrishnan, C.; Lakra, R.; Pardeshi, P.; Gupta, N.; Gangakhedkar, R.R.; et al. Nipah virus sequences from humans and bats during Nipah outbreak, Kerala, India, 2018. Emerg. Infect. Dis. 2019, 25, 1003–1006.
- AbuBakar, S.; Chang, L.Y.; Ali, A.R.; Sharifah, S.H.; Yusoff, K.; Zamrod, Z. Isolation and molecular identification of Nipah virus from pigs. Emerg. Infect. Dis. 2004, 10, 2228–2230.
- Clayton, B.A.; Middleton, D.; Arkinstall, R.; Frazer, L.; Wang, L.F.; Marsh, G.A. The nature of exposure drives transmission of Nipah viruses from Malaysia and Bangladesh in ferrets. PLoS Negl. Trop. Dis. 2016, 10, e0004775.
- Clayton, B.A.; Middleton, D.; Bergfeld, J.; Haining, J.; Arkinstall, R.; Wang, L.; Marsh, G.A. Transmission routes for Nipah virus from Malaysia and Bangladesh. Emerg. Infect. Dis. 2012, 18, 12–18.
- Mire, C.E.; Satterfield, B.A.; Geisbert, J.B.; Agans, K.N.; Borisevich, V.; Yan, L.; Chan, Y.P.; Cross, R.W.; Fenton, K.A.; Broder, C.C.; et al. Pathogenic differences between Nipah virus Bangladesh and Malaysia strains in primates: Implications for antibody therapy. Sci. Rep. 2016, 6, 30916.
- Pro-Med. Nipah Virus—Bangladesh, India; WHO: Geneva, Switzerland, 2021; p. 20210910.28660529.
- Chong, H.T.; Hossain, M.J.; Tan, C.T. Differences in epidemiologic and clinical features of Nipah virus encephalitis between the Malaysian and Bangladesh outbreaks. Neurol. Asia 2008, 13, 23–26.
- Chandni, R.; Renjith, T.P.; Fazal, A.; Yoosef, N.; Ashhar, C.; Thulaseedharan, N.K.; Suraj, K.P.; Sreejith, M.K.; Sajeeth Kumar, K.G.; Rajendran, V.R.; et al. Clinical manifestations of Nipah virus-infected patients who presented to the Emergency Department during an outbreak in Kerala state in India, May 2018. Clin. Infect. Dis. 2020, 71, 152–157.
- Rahmat, K.; Goh, K.J. Late-onset Nipah virus encephalitis 11 years after the initial outbreak: A case report. Neurol. Asia 2012, 17, 71–74.
- Tan, C.T.; Goh, K.J.; Wong, K.T.; Sarji, S.A.; Chua, K.B.; Chew, N.K.; Murugasu, P.; Loh, Y.L.; Chong, H.T.; Tan, K.S.; et al. Relapsed and late-onset Nipah encephalitis. Ann. Neurol. 2002, 51, 703–708.
- Chua, K.B.; Lek Koh, C.; Hooi, P.S.; Wee, K.F.; Khong, J.H.; Chua, B.H.; Chan, Y.P.; Lim, M.E.; Lam, S.K. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 2002, 4, 145–151.
- Halpin, K.; Hyatt, A.D.; Fogarty, R.; Middleton, D.; Bingham, J.; Epstein, J.H.; Rahman, S.A.; Hughes, T.; Smith, C.; Field, H.E.; et al. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: A comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg. 2011, 85, 946–951.
- Rahman, S.A.; Hassan, S.S.; Olival, K.J.; Mohamed, M.; Chang, L.Y.; Hassan, L.; Saad, N.M.; Shohaimi, S.A.; Mamat, Z.C.; Naim, M.S.; et al. Characterization of Nipah virus from naturally infected Pteropus vampyrus bats, Malaysia. Emerg. Infect. Dis. 2010, 16, 1990–1993.
- Islam, M.S.; Sazzad, H.M.S.; Satter, S.M.; Sultana, S.; Hossain, M.J.; Hasan, M.; Rahman, M.; Campbell, S.; Cannon, D.L.; Ströher, U.; et al. Nipah virus transmission from bats to humans associated with drinking traditional liquor made from date palm sap, Bangladesh, 2011–2014. Emerg. Infect. Dis. 2016, 22, 664–670.
- Homaira, N.; Rahman, M.; Hossain, M.J.; Epstein, J.H.; Sultana, R.; Khan, M.S.U.; Podder, G.; Nahar, K.; Ahmed, B.; Gurley, E.S.; et al. Nipah virus outbreak with person-to-person transmission in a district of Bangladesh, 2007. Epidemiol. Infect. 2010, 138, 1630–1636.
- Salah Uddin Khan, M.; Hossain, J.; Gurley, E.S.; Nahar, N.; Sultana, R.; Luby, S.P. Use of infrared camera to understand bats’ access to date palm sap: Implications for preventing Nipah virus transmission. Ecohealth 2010, 7, 517–525.
- Gurley, E.; Montgomery, J.; Hossain, M.; Bell, M.; Azad, A.; Islam, M.; Molla, M.; Carroll, D.; Ksiazek, T.; Rota, P.; et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg. Infect. Dis. 2007, 13, 1031–1037.
- Luby, S.P.; Hossain, M.J.; Gurley, E.S.; Ahmed, B.N.; Banu, S.; Khan, S.U.; Homaira, N.; Rota, P.A.; Rollin, P.E.; Comer, J.A.; et al. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg. Infect. Dis. 2009, 15, 1229–1235.
- Devnath, P.; Masud, H.M.A.A. Nipah virus: A potential pandemic agent in the context of the current severe acute respiratory syndrome coronavirus 2 pandemic. New Microbes New Infect. 2021, 41, 100873.
- Luby, S.P. The pandemic potential of Nipah virus. Antivir. Res. 2013, 100, 38–43.
- Gomez Roman, R.; Tornieporth, N.; Cherian, N.G.; Shurtleff, A.C.; L’Azou Jackson, M.; Yeskey, D.; Hacker, A.; Mungai, E.; Le, T.T. Medical countermeasures against henipaviruses: A review and public health perspective. Lancet Infect. Dis. 2022, 22, e13–e27.
- Bonaparte, M.I.; Dimitrov, A.S.; Bossart, K.N.; Crameri, G.; Mungall, B.A.; Bishop, K.A.; Choudhry, V.; Dimitrov, D.S.; Wang, L.F.; Eaton, B.T.; et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 2005, 102, 10652–10657.
- Xu, K.; Rajashankar, K.R.; Chan, Y.-P.; Himanen, J.P.; Broder, C.C.; Nikolov, D.B. Host cell recognition by the henipaviruses: Crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. USA 2008, 105, 9953–9958.
- Negrete, O.A.; Wolf, M.C.; Aguilar, H.C.; Enterlain, S.; Wang, W.; Mühlberger, E.; Su, S.V.; Bertolotti-Ciarlet, A.; Flick, R.; Lee, B. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006, 2, e7.
- Geisbert, T.W.; Daddario-DiCaprio, K.M.; Hickey, A.C.; Smith, M.A.; Chan, Y.-P.; Wang, L.-F.; Mattapallil, J.J.; Geisbert, J.B.; Bossart, K.N.; Broder, C.C. Development of an acute and highly pathogenic nonhuman primate model of Nipah virus infection. PLoS ONE 2010, 5, e10690.
- Guillaume, V.; Wong, K.T.; Looi, R.; Georges-Courbot, M.-C.; Barrot, L.; Buckland, R.; Wild, T.F.; Horvat, B. Acute Hendra virus infection: Analysis of the pathogenesis and passive antibody protection in the hamster model. Virology 2009, 387, 459–465.
- Bossart, K.N.; Tachedjian, M.; McEachern, J.A.; Crameri, G.; Zhu, Z.; Dimitrov, D.S.; Broder, C.C.; Wang, L.-F. Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology 2008, 372, 357–371.
- DeBuysscher, B.L.; Scott, D.P.; Rosenke, R.; Wahl, V.; Feldmann, H.; Prescott, J. Nipah virus efficiently replicates in human smooth muscle cells without cytopathic effect. Cells 2021, 10, 1319.
- Tiong, V.; Shu, M.-H.; Wong, W.F.; AbuBakar, S.; Chang, L.-Y. Nipah virus infection of immature dendritic cells increases its transendothelial migration across human brain microvascular endothelial cells. Front. Microbiol. 2018, 9, 2747.
- Liu, J.; Coffin, K.M.; Johnston, S.C.; Babka, A.M.; Bell, T.M.; Long, S.Y.; Honko, A.N.; Kuhn, J.H.; Zeng, X. Nipah virus persists in the brains of nonhuman primate survivors. JCI Insight 2019, 4, e129629.
- Wong, K.; Robertson, T.; Ong, B.; Chong, J.; Yaiw, K.; Wang, L.; Ansford, A.; Tannenberg, A. Human Hendra virus infection causes acute and relapsing encephalitis. Neuropathol. Appl. Neurobiol. 2009, 35, 296–305.
More
