Microphthalmia-associated transcription factor (MITF) is an important regulator of melanogenesis and melanocyte development. Cutaneous malignant melanomas are heterogeneous in nature, comprising several cell subpopulations with distinct transcriptomic signatures and behaviours. Melanomas carrying different genetic alterations have different clinical features and different relation with sun exposure. MITF-low cutaneous melanoma cells display a higher expression of stem cell markers (OCT4 and NANOG) and are able to produce larger tumours when injected into nude mice. However, both MITF-low and MITF-high cells can give rise to tumours, which then contain both types of cells. Uveal melanomas are malignant tumours that originate in the uveal tract of the eye and have different mutations and behaviour compared to cutaneous melanoma. The role of MITF in uveal melanoma is not clearly defined, but MITF loss is associated with loss of BAP1 expression, which is a marker of poor prognosis,
- melanoma
- MITF
- melanocyte
- cutaneous melanoma
- uveal melanoma
1. Cell Proliferation
2. Cell Survival
3. Epithelial-Mesenchymal Transition and Motile Ability
4. Regulation of MITF in Cutaneous Melanoma
5. Clinical Relevance
6. MITF in uveal melanoma
Uveal melanoma is the most common primary intraocular malignancy in adults. Despite originating from melanocytes as well, it is very different from cutaneous melanoma: it uveal melanoma does not have a UV signature, does not carry the typical cutaneous melanoma mutations and does not respond well to target or immunotherapy. A UM usually carries either a GNAQ or a GNA11 mutation [171,172]. When a second mutation occurs, it usually involves one of three genes: BAP1, EIF1AX, or SF3B1 [8,173]. A few studies analysed the presence and function of MITF in UM and, as for cutaneous melanoma, its role seems to be complex: some studies classify MITF as prooncogenic, and others mention its expression as a feature of low-risk tumours. Mouriaux et al. found a positive correlation between MITF staining and proliferative activity [175]. Hippo-YAP/TAZ also emerged as an important signalling pathway downstream of GNAQ/11 that controls UM cell proliferation. Downstream of this module lies PAX3, which controls MITF expression [176]. As such, YAP inhibition suppressed the growth of UM [177,178]. In contrast, MITF has been shown to increase p16 expression in UM, where CDKN2A mutations have rarely been described, supporting the idea that MITF can induce cell cycle arrest and behave as a tumour suppressor gene [64]. miRNAs and epigenetic mechanisms are involved in MITF regulation in UM cells as well: miR-137 through downregulation of MITF, MET, and CDK6 (cyclin dependent kinase 6) and miR-182 through inhibition of MITF, MET, BCL2, and cyclin D2, causing G1 cell cycle arrest, thereby reducing the number of metabolically-active UM cells [179,180]. Increase in miR-137 by the DNA hypomethylating agent 5-aza-20-deoxycytidineor the histone deacetylase inhibitor trichostatin A (TSA) also represents a therapeutic opportunity to impair UM cell proliferation through MITF inhibition [179]. However, as in cutaneous melanoma, MITF inhibition might be associated with a stem cell-like phenotype. Matatall et al. showed that BAP1 knockdown was associated with an increase in expression of stem cell markers (NANOG), and loss of pigmentation markers (MITF, TYR, and DCT) and with the capacity of UM cells to form anchorage-independent colonies. These data suggest that BAP1 loss induced a dedifferentiated and a stem-like phenotype in UM cells, although this remains to be fully demonstrated [181].
References
- King, R.; Googe, P.B.; Weilbaecher, K.N.; Mihm, M.C., Jr.; Fisher, D.E. Microphthalmia Transcription Factor Expression in Cutaneous Benign, Malignant Melanocytic, and Nonmelanocytic Tumors. Am. J. Surg. Pathol. 2001, 25, 51–57.
- Granter, S.R.; Weilbaecher, K.N.; Quigley, C.; Fletcher, C.D.; Fisher, D.E. Microphthalmia transcription factor: Not a sensitive or specific marker for the diagnosis of desmoplastic melanoma and spindle cell (non-desmoplastic) melanoma. Am. J. Dermatopathol. 2001, 23, 185–189.
- Garraway, L.A.; Widlund, H.R.; Rubin, M.A.; Getz, G.; Berger, A.J.; Ramaswamy, S.; Beroukhim, R.; Milner, J.D.A.; Granter, S.R.; Du, J.; et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005, 436, 117–122.
- Bertolotto, C.; Lesueur, F.; Giuliano, S.; Strub, T.; De Lichy, M.; Bille, K.; Dessen, P.; D’Hayer, B.; Mohamdi, H.; Remenieras, A.; et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011, 480, 94–98.
- Ainger, S.A.; Jagirdar, K.; Lee, K.J.; Soyer, H.P.; Sturm, R.A. Skin Pigmentation Genetics for the Clinic. Dermatology 2017, 233, 1–15.
- Wellbrock, C.; Arozarena, I. Microphthalmia-associated transcription factor in melanoma development and MAP-kinase pathway targeted therapy. Pigment. Cell Melanoma Res. 2015, 28, 390–406.
- Rodriguez-Teja, M.; Aladowicz, E.; Lanfrancone, L.; Goding, C.R. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res. 2008, 68, 7872–7881.
- Carreira, S.; Liu, B.; Goding, C.R. The gene encoding the T-box factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes. J. Biol. Chem. 2000, 275, 21920–21927.
- Jacobs, J.J.L.; Keblusek, P.; Robanus-Maandag, E.; Kristel, P.; Lingbeek, M.; Nederlof, P.M.; Van Welsem, T.; van de Vijver, M.J.; Koh, E.Y.; Daley, G.Q.; et al. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19ARF) and is amplified in a subset of human breast cancers. Nat. Genet. 2000, 26, 291–299.
- Prince, S.; Carreira, S.; Vance, K.W.; Abrahams, A.; Goding, C.R. Tbx2 Directly Represses the Expression of the p21WAF1Cyclin-Dependent Kinase Inhibitor. Cancer Res. 2004, 64, 1669–1674.
- Strub, T.; Giuliano, S.; Ye, T.; Bonet, C.; Keime, C.; Kobi, D.; Le Gras, S.; Cormont, M.; Ballotti, R.; Bertolotto, C.; et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 2011, 30, 2319–2332.
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; LaRue, L.; Goding, C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006, 20, 3426–3439.
- Ploper, D.; Taelman, V.F.; Robert, L.; Perez, B.S.; Titz, B.; Chen, H.-W.; Graeber, T.G.; von Euw, E.; Ribas, A.; De Robertis, E.M. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. USA 2015, 112, E420–E429.
- Zoncu, R.; Bar-Peled, L.; Efeyan, A.; Wang, S.; Sancak, Y.; Sabatini, D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011, 334, 678–683.
- Haq, R.; Shoag, J.; Andreu-Perez, P.; Yokoyama, S.; Edelman, H.; Rowe, G.C.; Frederick, D.T.; Hurley, A.D.; Nellore, A.; Kung, A.L.; et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 2013, 23, 302–315.
- Vazquez, F.; Lim, J.-H.; Chim, H.; Bhalla, K.; Girnun, G.; Pierce, K.; Clish, C.B.; Granter, S.R.; Widlund, H.R.; Spiegelman, B.M.; et al. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 2013, 23, 287–301.
- Giuliano, S.; Cheli, Y.; Ohanna, M.; Bonet, C.; Beuret, L.; Bille, K.; Loubat, A.; Hofman, V.; Hofman, P.; Ponzio, G.; et al. Microphthalmia-Associated Transcription Factor Controls the DNA Damage Response and a Lineage-Specific Senescence Program in Melanomas. Cancer Res. 2010, 70, 3813–3822.
- Carreira, S.; Goodall, J.; Aksan, I.; La Rocca, S.A.; Galibert, M.-D.; Denat, L.; Larue, L.; Goding, C.R. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 2005, 433, 764–769.
- Haq, R.; Yokoyama, S.; Hawryluk, E.B.; Jönsson, G.B.; Frederick, D.T.; McHenry, K.; Porter, D.; Tran, T.-N.; Love, K.T.; Langer, R.; et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc. Natl. Acad. Sci. USA 2013, 110, 4321–4326.
- McGill, G.G.; Horstmann, M.A.; Widlund, H.; Du, J.; Motyckova, G.; Nishimura, E.K.; Lin, Y.-L.; Ramaswamy, S.; Avery, W.; Ding, H.-F.; et al. Bcl2 Regulation by the Melanocyte Master Regulator Mitf Modulates Lineage Survival and Melanoma Cell Viability. Cell 2002, 109, 707–718.
- Dynek, J.N.; Chan, S.M.; Liu, J.; Zha, J.; Fairbrother, W.J.; Vucic, D. Microphthalmia-Associated Transcription Factor Is a Critical Transcriptional Regulator of Melanoma Inhibitor of Apoptosis in Melanomas. Cancer Res. 2008, 68, 3124–3132.
- Beuret, L.; Flori, E.; Denoyelle, C.; Bille, K.; Busca, R.; Picardo, M.; Bertolotto, C.; Ballotti, R. Up-regulation of MET Expression by α-Melanocyte-stimulating Hormone and MITF Allows Hepatocyte Growth Factor to Protect Melanocytes and Melanoma Cells from Apoptosis. J. Biol. Chem. 2007, 282, 14140–14147.
- Liu, F.; Fu, Y.; Meyskens, F.L., Jr. MiTF Regulates Cellular Response to Reactive Oxygen Species through Transcriptional Regulation of APE-1/Ref-1. J. Investig. Dermatol. 2009, 129, 422–431.
- Logsdon, D.P.; Grimard, M.; Luo, M.; Shahda, S.; Jiang, Y.; Tong, Y.; Yu, Z.; Zyromski, N.; Schipani, E.; Carta, F.; et al. Regulation of HIF1α under Hypoxia by APE1/Ref-1 Impacts CA9 Expression: Dual Targeting in Patient-Derived 3D Pancreatic Cancer Models. Mol. Cancer Ther. 2016, 15, 2722–2732.
- Buscà, R.; Berra, E.; Gaggioli, C.; Khaled, M.; Bille, K.; Marchetti, B.; Thyss, R.; Fitsialos, G.; Larribère, L.; Bertolotto, C.; et al. Hypoxia-inducible factor 1α is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells. J. Cell Biol. 2005, 170, 49–59.
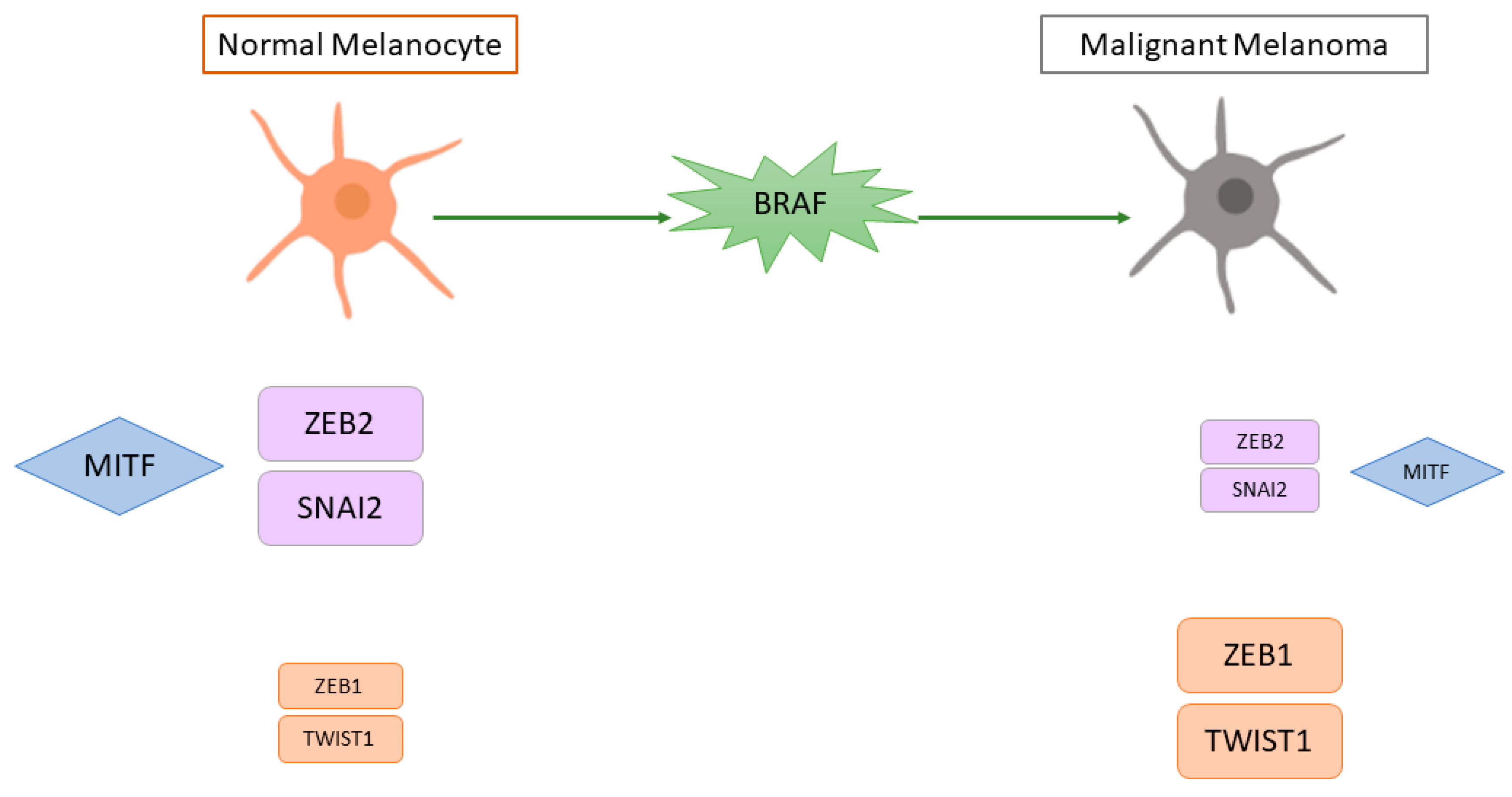
- Caramel, J.; Papadogeorgakis, E.; Hill, L.; Browne, G.J.; Richard, G.; Wierinckx, A.; Saldanha, G.; Osborne, J.; Hutchinson, P.; Tse, G.; et al. A Switch in the Expression of Embryonic EMT-Inducers Drives the Development of Malignant Melanoma. Cancer Cell 2013, 24, 466–480.
- Denecker, G.; Vandamme, N.; Akay, O.; Koludrovic, D.; Taminau, J.; Lemeire, K.; Gheldof, A.; De Craene, B.; Van Gele, M.; Brochez, L.; et al. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 2014, 21, 1250–1261.
- Vandamme, N.; Denecker, G.; Bruneel, K.; Blancke, G.; Akay, O.; Taminau, J.; De Coninck, J.; De Smedt, E.; Skrypek, N.; Van Loocke, W.; et al. The EMT Transcription Factor ZEB2 Promotes Proliferation of Primary and Metastatic Melanoma While Suppressing an Invasive, Mesenchymal-Like Phenotype. Cancer Res. 2020, 80, 2983–2995.
- Naffouje, S.; Naffouje, R.; Bhagwandin, S.; Salti, G.I. Microphthalmia transcription factor in malignant melanoma predicts occult sentinel lymph node metastases and survival. Melanoma Res. 2015, 25, 496–502.
- Salti, G.I.; Manougian, T.; Farolan, M.; Shilkaitis, A.; Majumdar, D.; Das Gupta, T.K. Micropthalmia transcription factor: A new prognostic marker in intermediate-thickness cutaneous malignant melanoma. Cancer Res. 2000, 60, 5012–5016.
- Goding, C.R.; Arnheiter, H. MITF—The first 25 years. Genes Dev. 2019, 33, 983–1007.
- Falletta, P.; del Campo, L.S.; Chauhan, J.; Effern, M.; Kenyon, A.; Kershaw, C.J.; Siddaway, R.; Lisle, R.J.; Freter, R.; Daniels, M.J.; et al. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 2017, 31, 18–33.
- Landsberg, J.; Kohlmeyer, J.; Renn, M.; Bald, T.; Rogava, M.; Cron, M.; Fatho, M.; Lennerz, V.; Wölfel, T.; Hölzel, M.; et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 2012, 490, 412–416.
- Riesenberg, S.; Groetchen, A.; Siddaway, R.; Bald, T.; Reinhardt, J.; Smorra, D.; Kohlmeyer, J.; Renn, M.; Phung, B.; Aymans, P.; et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat. Commun. 2015, 6, 8755.
- Goodall, J.; Carreira, S.; Denat, L.; Kobi, D.; Davidson, I.; Nuciforo, P.; Sturm, R.A.; LaRue, L.; Goding, C.R. Brn-2 represses microphthalmia-associated transcription factor expression and marks a distinct subpopulation of microphthalmia-associated transcription factor-negative melanoma cells. Cancer Res. 2008, 68, 7788–7794.
- Boyle, G.M.; Woods, S.L.; Bonazzi, V.F.; Stark, M.S.; Hacker, E.; Aoude, L.G.; Dutton-Regester, K.; Cook, A.L.; Sturm, R.A.; Hayward, N.K. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011, 24, 525–537.
- Segura, M.F.; Hanniford, D.; Menendez, S.; Reavie, L.; Zou, X.; Alvarez-Diaz, S.; Zakrzewski, J.; Blochin, E.; Rose, A.; Bogunovic, D.; et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. USA 2009, 106, 1814–1819.
- Cheli, Y.; Giuliano, S.; Fenouille, N.; Allegra, M.; Hofman, V.; Hofman, P.; Bahadoran, P.; Lacour, J.-P.; Tartare-Deckert, S.; Bertolotto, C.; et al. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene 2012, 31, 2461–2470.
- Vivas-García, Y.; Falletta, P.; Liebing, J.; Louphrasitthiphol, P.; Feng, Y.; Chauhan, J.; Scott, D.A.; Glodde, N.; Calvo, A.C.; Bonham, S.; et al. Lineage-Restricted Regulation of SCD and Fatty Acid Saturation by MITF Controls Melanoma Phenotypic Plasticity. Mol. Cell 2019, 77, 120–137.e9.
- Feige, E.; Yokoyama, S.; Levy, C.; Khaled, M.; Igras, V.; Lin, R.J.; Lee, S.; Widlund, H.R.; Granter, S.R.; Kung, A.L.; et al. Hypoxia-induced transcriptional repression of the melanoma-associated oncogene MITF. Proc. Natl. Acad. Sci. USA 2011, 108, E924–E933.
- Slominski, A.; Zbytek, B.; Slominski, R. Inhibitors of melanogenesis increase toxicity of cyclophosphamide and lymphocytes against melanoma cells. Int. J. Cancer 2009, 124, 1470–1477.
- Slominski, A.; Kim, T.-K.; Brożyna, A.; Janjetovic, Z.; Brooks, D.; Schwab, L.; Skobowiat, C.; Jóźwicki, W.; Seagroves, T. The role of melanogenesis in regulation of melanoma behavior: Melanogenesis leads to stimulation of HIF-1α expression and HIF-dependent attendant pathways. Arch. Biochem. Biophys. 2014, 563, 79–93.
- Martínez-Esparza, M.; Jiménez-Cervantes, C.; Beermann, F.; Aparicio, P.; Lozano, J.A.; Garcia-Borron, J.C. Transforming Growth Factor-β1 Inhibits Basal Melanogenesis in B16/F10 Mouse Melanoma Cells by Increasing the Rate of Degradation of Tyrosinase and Tyrosinase-related Protein-1. J. Biol. Chem. 1997, 272, 3967–3972.
- Hoek, K.S.; Schlegel, N.C.; Brafford, P.; Sucker, A.; Ugurel, S.; Kumar, R.; Weber, B.L.; Nathanson, K.L.; Phillips, D.J.; Herlyn, M.; et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006, 19, 290–302.
- Cheli, Y.; Guiliano, S.; Botton, T.; Rocchi, S.; Hofman, V.; Hofman, P.; Bahadoran, P.; Bertolotto, C.; Ballotti, R. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene 2011, 30, 2307–2318.
- Rambow, F.; Marine, J.-C.; Goding, C.R. Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 2019, 33, 1295–1318.
- Smith, M.P.; Sanchez-Laorden, B.; O’Brien, K.; Brunton, H.; Ferguson, J.; Young, H.; Dhomen, N.; Flaherty, K.T.; Frederick, D.T.; Cooper, Z.A.; et al. The immune microenvironment confers resistance to MAPK pathway inhibitors through macrophage-derived TNFalpha. Cancer Discov. 2014, 4, 1214–1229.
- Ohanna, M.; Giuliano, S.; Bonet, C.; Imbert, V.; Hofman, V.; Zangari, J.; Bille, K.; Robert, C.; Paillerets, B.B.-D.; Hofman, P.; et al. Senescent cells develop a PARP-1 and nuclear factor-κB-associated secretome (PNAS). Genes Dev. 2011, 25, 1245–1261.
- Ohanna, M.; Cheli, Y.; Bonet, C.; Bonazzi, V.F.; Allegra, M.; Giuliano, S.; Bille, K.; Bahadoran, P.; Giacchero, D.; Lacour, J.-P.; et al. Secretome from senescent melanoma engages the STAT3 pathway to favor reprogramming of naive melanoma towards a tumor-initiating cell phenotype. Oncotarget 2013, 4, 2212–2224.
- Konieczkowski, D.J.; Johannessen, C.M.; Abudayyeh, O.; Kim, J.W.; Cooper, Z.A.; Piris, A.; Frederick, D.T.; Barzily-Rokni, M.; Straussman, R.; Haq, R.; et al. A Melanoma Cell State Distinction Influences Sensitivity to MAPK Pathway Inhibitors. Cancer Discov. 2014, 4, 816–827.
- Estrada, C.; Mirabal-Ortega, L.; Méry, L.; Dingli, F.; Besse, L.; Messaoudi, C.; Loew, D.; Pouponnot, C.; Bertolotto, C.; Eychène, A.; et al. MITF activity is regulated by a direct interaction with RAF proteins in melanoma cells. Commun. Biol. 2022, 5, 101.
- Brozyna, A.A.; Jóźwicki, W.; Carlson, J.A.; Slominski, A.T. Melanogenesis affects overall and disease-free survival in patients with stage III and IV melanoma. Hum. Pathol. 2013, 44, 2071–2074.
- Brożyna, A.A.; Jóźwicki, W.; Roszkowski, K.; Filipiak, J.; Slominski, A.T. Melanin content in melanoma metastases affects the outcome of radiotherapy. Oncotarget 2016, 7, 17844–17853.
- Slominski, R.M.; Sarna, T.; Płonka, P.M.; Raman, C.; Brożyna, A.A.; Slominski, A.T. Melanoma, Melanin, and Melanogenesis: The Yin and Yang Relationship. Front. Oncol. 2022, 12, 842496.
- Ballotti, R.; Cheli, Y.; Bertolotto, C. The complex relationship between MITF and the immune system: A Melanoma ImmunoTherapy (response) Factor? Mol. Cancer 2020, 19, 170.
- Arts, N.; Cané, S.; Hennequart, M.; Lamy, J.; Bommer, G.; Van Den Eynde, B.; De Plaen, E. microRNA-155, induced by interleukin-1ss, represses the expression of microphthalmia-associated transcription factor (MITF-M) in melanoma cells. PLoS ONE 2015, 10, e0122517.
- Müller, J.; Krijgsman, O.; Tsoi, J.; Robert, L.; Hugo, W.; Song, C.; Kong, X.; Possik, P.A.; Cornelissen-Steijger, P.D.; Foppen, M.H.G.; et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 2014, 5, 5712.
- Van Raamsdonk, C.D.; Bezrookove, V.; Green, G.; Bauer, J.; Gaugler, L.; O’Brien, J.M.; Simpson, E.M.; Barsh, G.S.; Bastian, B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009, 457, 599–602.
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in GNA11 in Uveal Melanoma. N. Engl. J. Med. 2010, 363, 2191–2199.
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.-H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A.; et al. Uveal melanoma. Nat. Rev. Dis. Prim. 2020, 6, 24.
- Smit, K.N.; Jager, M.J.; de Klein, A.; Kiliҫ, E. Uveal melanoma: Towards a molecular understanding. Prog. Retin. Eye Res. 2020, 75, 100800.
- Mouriaux, F.; Vincent, S.; Kherrouche, Z.; Maurage, C.-A.; Planque, N.; Monté, D.; Labalette, P.; Saule, S. Microphthalmia transcription factor analysis in posterior uveal melanomas. Exp. Eye Res. 2003, 76, 653–661.
- Manderfield, L.J.; Engleka, K.A.; Aghajanian, H.; Gupta, M.; Yang, S.; Li, L.; Baggs, J.E.; Hogenesch, J.B.; Olson, E.N.; Epstein, J.A. Pax3 and Hippo Signaling Coordinate Melanocyte Gene Expression in Neural Crest. Cell Rep. 2014, 9, 1885–1895.
- Lyubasyuk, V.; Ouyang, H.; Yu, F.-X.; Guan, K.-L.; Zhang, K. YAP inhibition blocks uveal melanogenesis driven by GNAQ or GNA11 mutations. Mol. Cell. Oncol. 2015, 2, e970957.
- Brouwer, N.J.; Konstantinou, E.K.; Gragoudas, E.S.; Marinkovic, M.; Luyten, G.P.M.; Kim, I.K.; Jager, M.J.; Vavvas, D.G. Targeting the YAP/TAZ Pathway in Uveal and Conjunctival Melanoma with Verteporfin. Investig. Opthalmol. Vis. Sci. 2021, 62, 3.
- Loercher, A.E.; Tank, E.M.; Delston, R.B.; Harbour, J.W. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J. Cell Biol. 2005, 168, 35–40.
- Chen, X.; Wang, J.; Shen, H.; Lu, J.; Li, C.; Hu, D.-N.; Dong, X.D.; Yan, D.; Tu, L. Epigenetics, MicroRNAs, and Carcinogenesis: Functional Role of MicroRNA-137 in Uveal Melanoma. Investig. Ophthalmol. Vis. Sci. 2011, 52, 1193–1199.
- Yan, D.; Dong, X.D.; Chen, X.; Yao, S.; Wang, L.; Wang, J.; Wang, C.; Hu, D.-N.; Qu, J.; Tu, L. Role of MicroRNA-182 in Posterior Uveal Melanoma: Regulation of Tumor Development through MITF, BCL2 and Cyclin D2. PLoS ONE 2012, 7, e40967.
- Matatall, K.A.; Agapova, O.A.; Onken, M.D.; Worley, L.A.; Bowcock, A.M.; Harbour, J.W. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer 2013, 13, 371.
