Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Vivi Li and Version 1 by Shanshan Xie.
N1-methyladenosine (m1A) is a prevalent and reversible post-transcriptional RNA modification that decorates tRNA, rRNA and mRNA. Recent sStudies based on technical advances in analytical chemistry and high-throughput sequencing methods have revealed the crucial roles of m1A RNA modification in gene regulation and biological processes.
- N1-methyladenosine(m1A)
- RNA modification
- gene expression
1. Introduction
Cellular RNAs contain more than 170 different types of chemical modifications across species [1]. N1-methyladenosine(m1A) is a reversible methylation involving the addition of a methyl group at the N1 position of adenosine in cellular transcripts [2]. The methyl group can block the normal Watson–Crick base pairing of A:T or A:U, resulting in an unstable mismatch with other nucleosides by forming Hoogsteen base pairs [3]. The secondary structure and RNA–protein interaction of m1A-modified RNAs are also altered under physiological conditions [4]. As a dynamic and reversible post-transcriptional RNA modification, m1A can be installed by methyltransferases, removed by demethylases and recognized by m1A-dependent RNA-binding proteins [2,5][2][5]. m1A RNA modification affects RNA metabolism, including RNA structure, stability and mRNA translation, thereby regulating gene expression and several fundamental cellular processes [6].
m1A RNA modification has been found with high abundance in transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) but at low levels in messenger RNAs (mRNAs) [7,8,9,10,11,12][7][8][9][10][11][12]. It occurs in the tRNA of bacteria, archaea and eukaryotes at positions 9, 14, 16, 22, 57 and 58 (m1A9, m1A14, m1A16, m1A22, m1A57, and m1A58, respectively) [13]. In cytosolic (cyt) tRNAs, m1A RNA modification occurs at five different positions (9, 14, 22, 57, and 58) [14,15][14][15]. Among them, m1A14 has only been identified in cyt(tRNA)Phe from mammals, m1A22 has only been identified in bacteria tRNAs, and m1A57 has been identified in archaea existing only transiently as an intermediate of 1-methylinosine (m1I) [14,15][14][15]. In mitochondria, m1A9 is quite abundant and found in 14 species of mt-tRNA, while m1A58 is a minor modification with a 17% frequency found in four species of mt-tRNAs [16]. Additionally, m1A16 is unique to human mt-tRNAArg, and its frequency is approximately 20% [16]. For rRNAs, the nuclear-encoded large subunit rRNA m1A645 in 25S rRNA and m1A1322 in 28S rRNA located in the peptidyl transfer center of the ribosome are conserved in budding yeast and humans, respectively [17,18[17][18][19],19], and m1A is conserved at position 947 of 16S rRNA in the mitochondrial ribosome of vertebrates [20]. Regarding mRNAs, m1A in mRNA accounts for approximately 0.015–0.054% of all adenosines in mammalian cell lines and 0.05–0.16% in mammalian tissues [9,10,21][9][10][21]. m1A sites are usually located near the translation start site and the first splice site of mRNA, and they are associated with the translation of coding transcripts [9,10][9][10].
2. m1A RNA-Modifying Proteins
Reversible m1A methylomes in nuclear- and mitochondrial-encoded transcripts are achieved via the dynamic regulation of m1A RNA-modifying proteins (m1A methyltransferases, m1A demethylases and m1A-dependent RNA-binding proteins). The characterization of m1A-modifying proteins is crucial for understanding the mechanisms underlying m1A-mediated gene regulation and the biological roles of m1A RNA modification. To date, several m1A RNA-modifying proteins responsible for nuclear- and mitochondrial-encoded transcripts have been identified in humans (Figure 1).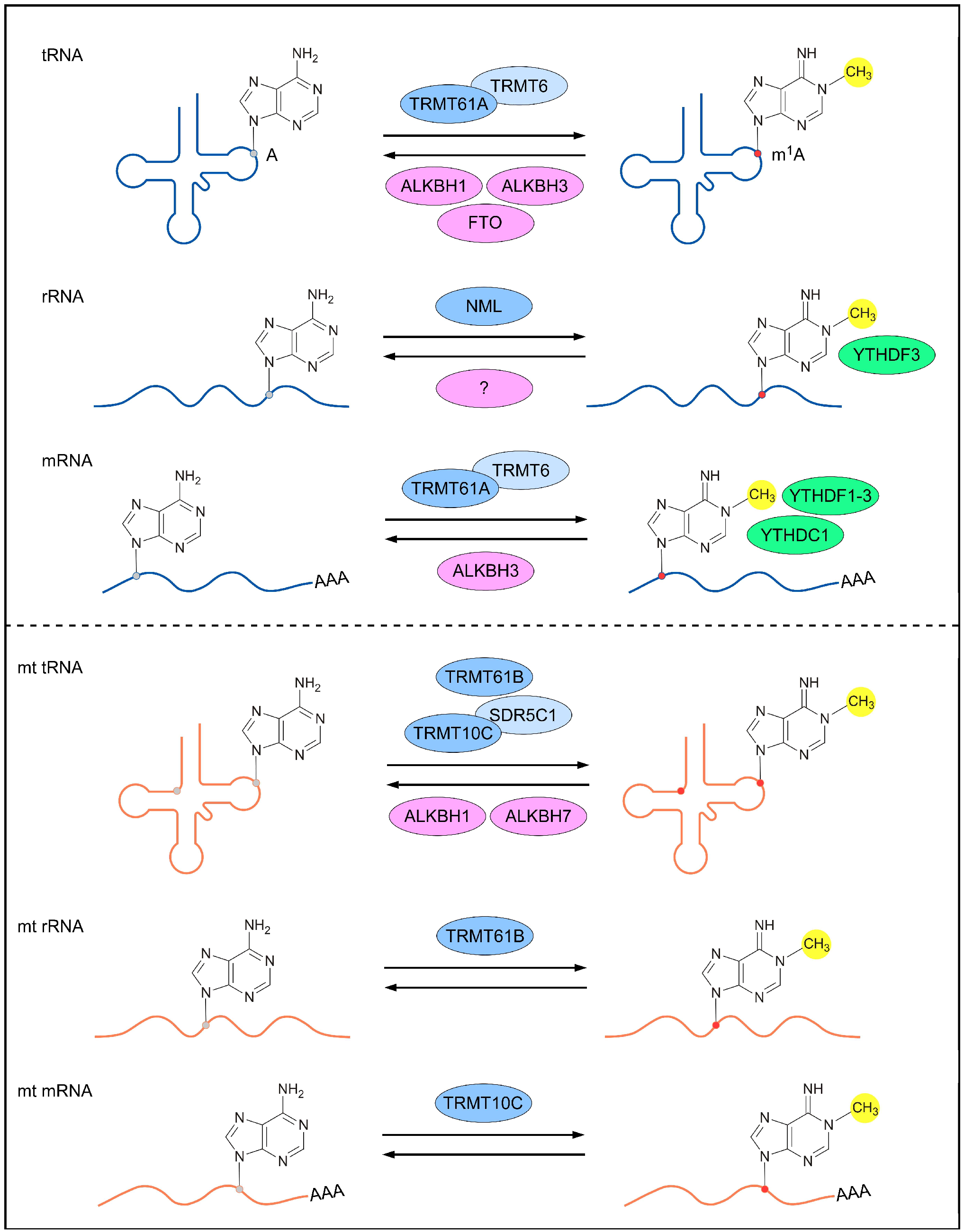
Figure 1. m1A-modifying proteins for different types of RNAs. The nuclear-encoded (top panel) and mitochondrial (bottom panel) RNAs are reversibly methylated by m1A methyltransferases (blue; dark blue represents catalytic core of the methylase complex), demethylased by m1A demethylases (pink), and bound by m1A-dependent RNA-binding proteins (green). A, adenosine; m1A, N1-methyladenosine; TRMT, tRNA (adenine (58)-N (1))-methyltransferase subunit; ALKBH, α-ketoglutarate-dependent dioxygenase alkB homolog; FTO, α-ketoglutarate-dependent dioxygenase alkB homolog FTO; NML, nucleomethylin; YTHDF, YTH domain-containing family protein; YTHDC1, YTH domain-containing protein 1; SDR5C1, 3-hydroxyacyl-CoA dehydrogenase type-2.
3. Biological Functions of m1A RNA Modification
Since the discovery of m1A RNA modification as a chemical modification of RNAs, efforts have been taken to understand the functional characterization of this dynamic methylation in RNA metabolism and gene expression regulation.3.1. m
1
A RNA Modification in RNA Metabolism
m1A RNA modification is a pivotal regulator of RNA metabolism, including RNA structure alteration, decay and translation (Figure 2).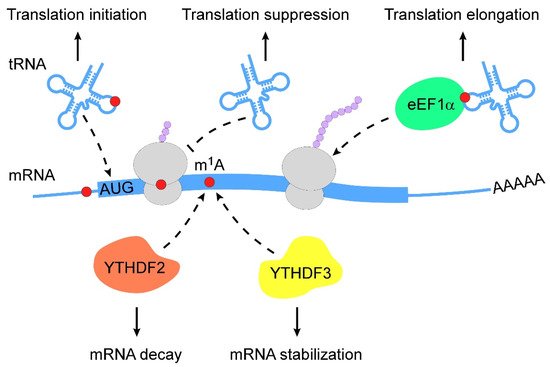
Figure 2. Action mechanisms of m1A in RNA metabolism. m1A RNA modification regulates RNA metabolism in multiple layers (from top to bottom: (1) m1A RNA modification stabilizes tRNAs to promote translation initiation; (2) m1A-modified mRNAs interfere with Watson–Crick base-pairing with tRNA to suppress translation; (3) m1A-modified tRNAs are coupled with eEF1α to polysomes to promote translation elongation; (4) m1A-modified mRNAs are subjected to degradation by interacting with YTHDF2; (5) m1A-modified mRNAs become stable when they bind to YTHDF3). m1A, N1-methyladenosine; eEF1α, eukaryotic elongation factor 1-α; YTHDF, YTH domain-containing family protein.
3.2. m
1
A RNA Modification in Biological Processes
Post-transcriptional modifications are involved in various biological processes, and recent evidence showed the importance of m1A RNA modification in this field. In a high-temperature-sensitive Thermococcus kodakarensis strain, decreased m1A58 and melting temperature of tRNA were observed, suggesting the relevance of m1A58 and the growth ability of this strain at high temperatures [49][34]. m1A RNA modification was found to exhibit its protective ability of RNAs under stress conditions. During heat shock, m1A-harbouring transcripts were found to preferentially accumulate in stress granules, subsequently resulting in a shorter time to restore the translation state during recovery [50][35]. Alkylating agents induced m1A modification in RNAs and orchestrated translational suppression by recruiting the ASCC damage repair complex (activating signal cointegrator 1 complex) [51][36]. The tRNA modification profiles of the Aplysia central nervous system showed increased m1A RNA modification levels in animals after behavioral training [52][37]; this was the first study to characterize the variable pattern of m1A RNA modification during defensive reflex-associated behavioral sensitization. Petunia TRMT61A catalyzed m1A RNA modification in mRNAs, and the knockdown of TRMT61A decreased the chlorophyll content and changed chlorotic and wrinkled leaf phenotype [53][38]. A recent study showed that the m1A demethylase ALKBH3 functioned as a negative regulator of ciliogenesis by removing the m1A sites on Aurora A mRNA (a key regulator of cilia disassembly) in mammalian cells, which was further involved in cilia-associated developmental processes in zebrafish [54][39].4. m1A RNA Modification in Diseases
The limited exploration of m1A RNA modification as a pathological feature has mainly focused on tumor progression (Table 1). It was reported that the knockdown of m1A demethylase ALKBH3 increased the abundance of m1A RNA modification in small RNAs (< 200 nucleotides) along with suppressed nascent protein in pancreatic cancer cells [55][40]. The ALKBH3-dependent m1A demethylation of macrophage colony-stimulating factor 1 (CSF1) mRNA enhanced its mRNA stability and thus promoted the invasion of breast and ovarian cancer cells [56][41]. In addition, ALKBH3 removed the m1A RNA modification of tRNAGlyGCC to promote tRNA cleavage by angiogenin. The generation of excessive tRNA-derived small RNAs may affect ribosome assembly and apoptosis in HeLa cells [57][42]. Furthermore, ALKBH3 promoter CpG island hypermethylation and transcriptional silencing were found in Hodgkin lymphoma cells, which were identified as a potential prognostic biomarker associated with poor clinical outcomes in patients with Hodgkin lymphoma [58][43]. A recent study found that levels of tRNA m1A modification were upregulated in hepatocellular carcinoma (HCC) tissues. The TRMT6/TRMT61A complex mediated increased m1A58 levels in tRNA, which then triggered peroxisome proliferator-activated receptor delta (PPARδ) mRNA translation in HCC stem cells. PPARδ promoted cholesterol biogenesis to activate the Hedgehog pathway, thereby initiating the self-renewal of HCC stem cells [59][44].Table 1.
Dysregulation of m
1
A RNA modification in human cancers.
| Cancers | m1A-Modifying Proteins | Roles | Targets | Mechanisms | Refs |
|---|---|---|---|---|---|
| Pancreatic cancer | ALKBH3 | Oncogene | small RNAs | Unknown | [55][40] |
| Breast and ovarian cancer | ALKBH3 | Oncogene | CSF1 | mRNA decay | [56][41] |
| Cervical cancer | ALKBH3 | Oncogene | tRNAs | tRNA cleavage | [57][42] |
| Hodgkin lymphoma | ALKBH3 | Tumor suppressor |
COL1A1, COL1A2 | Unknown | [58][43] |
| Hepatocellular carcinoma | TRMT6/TRMT61A | Oncogene | tRNAs | Unknown | [59][44] |
ALKBH, α-ketoglutarate-dependent dioxygenase alkB homolog; TRMT, tRNA (adenine(58)-N(1))-methyltransferase subunit; CSF-1, macrophage colony-stimulating factor 1; COL1A1, collagen α-1(I) chain; COL1A2, collagen α-2(I) chain.
References
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–D235.
- Wiener, D.; Schwartz, S. The epitranscriptome beyond m6A. Nat. Rev. Genet. 2021, 22, 119–131.
- Zhou, H.; Kimsey, I.J.; Nikolova, E.N.; Sathyamoorthy, B.; Grazioli, G.; McSally, J.; Bai, T.; Wunderlich, C.H.; Kreutz, C.; Andricioaei, I.; et al. m1A and m1G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat. Struct. Mol. Biol. 2016, 23, 803–810.
- Xiong, X.; Li, X.; Yi, C. N1-methyladenosine methylome in messenger RNA and non-coding RNA. Curr. Opin. Chem. Biol. 2018, 45, 179–186.
- Xu, G.L.; Bochtler, M. Reversal of nucleobase methylation by dioxygenases. Nat. Chem. Biol. 2020, 16, 1160–1169.
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42.
- Cozen, A.E.; Quartley, E.; Holmes, A.D.; Hrabeta-Robinson, E.; Phizicky, E.M.; Lowe, T.M. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 2015, 12, 879–884.
- Li, X.; Xiong, X.; Zhang, M.; Wang, K.; Chen, Y.; Zhou, J.; Mao, Y.; Lv, J.; Yi, D.; Chen, X.W.; et al. Base-Resolution Mapping Reveals Distinct m1A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol. Cell 2017, 68, 993–1005.
- Li, X.; Xiong, X.; Wang, K.; Wang, L.; Shu, X.; Ma, S.; Yi, C. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 2016, 12, 311–316.
- Dominissini, D.; Nachtergaele, S.; Moshitch-Moshkovitz, S.; Peer, E.; Kol, N.; Ben-Haim, M.S.; Dai, Q.; Di Segni, A.; Salmon-Divon, M.; Clark, W.C.; et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016, 530, 441–446.
- Zhou, H.; Rauch, S.; Dai, Q.; Cui, X.; Zhang, Z.; Nachtergaele, S.; Sepich, C.; He, C.; Dickinson, B.C. Evolution of a reverse transcriptase to map N1-methyladenosine in human messenger RNA. Nat. Methods 2019, 16, 1281–1288.
- Safra, M.; Sas-Chen, A.; Nir, R.; Winkler, R.; Nachshon, A.; Bar-Yaacov, D.; Erlacher, M.; Rossmanith, W.; Stern-Ginossar, N.; Schwartz, S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 2017, 551, 251–255.
- Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392.
- Oerum, S.; Dégut, C.; Barraud, P.; Tisné, C. m1A post-transcriptional modification in tRNAs. Biomolecules 2017, 7, 20.
- Motorin, Y.; Helm, M. RNA nucleotide methylation: 2021 update. Wiley Interdiscip. Rev. RNA 2022, 13, e1691.
- Suzuki, T.; Yashiro, Y.; Kikuchi, I.; Ishigami, Y.; Saito, H.; Matsuzawa, I.; Okada, S.; Mito, M.; Iwasaki, S.; Ma, D.; et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020, 11, 4269.
- Sharma, S.; Lafontaine, D.L.J. ‘View from a bridge’: A new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015, 40, 560–575.
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152.
- Sergiev, P.V.; Aleksashin, N.A.; Chugunova, A.A.; Polikanov, Y.S.; Dontsova, O.A. Structural and evolutionary insights into ribosomal RNA methylation. Nat. Chem. Biol. 2018, 14, 226–235.
- Bar-Yaacov, D.; Frumkin, I.; Yashiro, Y.; Chujo, T.; Ishigami, Y.; Chemla, Y.; Blumberg, A.; Schlesinger, O.; Bieri, P.; Greber, B.; et al. Mitochondrial 16S rRNA is methylated by tRNA methyltransferase TRMT61B in all vertebrates. PLoS Biol. 2016, 14, e1002557.
- Legrand, C.; Tuorto, F.; Hartmann, M.; Liebers, R.; Jacob, D.; Helm, M.; Lyko, F. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017, 27, 1589–1596.
- Helm, M.; Giegé, R.; Florentz, C. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 1999, 38, 13338–13346.
- Voigts-Hoffmann, F.; Hengesbach, M.; Kobitski, A.Y.; Aerschot, A.V.; Herdewijn, P.; Nienhaus, G.U.; Helm, M. A methyl group controls conformational equilibrium in Human mitochondrial tRNALys. J. Am. Chem. Soc. 2007, 129, 13382–13383.
- Wang, X.; Jia, H.; Jankowsky, E.; Anderson, J.T. Degradation of hypomodified tRNAiMet in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA 2008, 14, 107–116.
- Richter, U.; Evans, M.E.; Clark, W.C.; Marttinen, P.; Shoubridge, E.A.; Suomalainen, A.; Wredenberg, A.; Wedell, A.; Pan, T.; Battersby, B.J. RNA modification landscape of the human mitochondrial tRNALys regulates protein synthesis. Nat. Commun. 2018, 9, 3966.
- Sharma, S.; Hartmann, J.D.; Watzinger, P.; Klepper, A.; Peifer, C.; Kotter, P.; Lafontaine, D.L.J.; Entian, K.D. A single N1-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci. Rep. 2018, 8, 11904.
- Yang, T.; Cheong, A.; Mai, X.; Zou, S.; Woon, E.C. A methylation-switchable conformational probe for the sensitive and selective detection of RNA demethylase activity. Chem. Commun. 2016, 52, 6181–6184.
- Seo, K.W.; Kleiner, R.E. YTHDF2 recognition of N1-methyladenosine (m1A)-modified RNA is associated with transcript destabilization. ACS Chem. Biol. 2020, 15, 132–139.
- Zheng, Q.; Gan, H.; Yang, F.; Yao, Y.; Hao, F.; Hong, L.; Jin, L. Cytoplasmic m1A reader YTHDF3 inhibits trophoblast invasion by downregulation of m1A-methylated IGF1R. Cell Discov. 2020, 6, 12.
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell 2016, 167, 816–828.
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 2018, 71, 973–985.
- Fukuda, H.; Chujo, T.; Wei, F.Y.; Shi, S.L.; Hirayama, M.; Kaitsuka, T.; Yamamoto, T.; Oshiumi, H.; Tomizawa, K. Cooperative methylation of human tRNA3Lys at positions A58 and U54 drives the early and late steps of HIV-1 replication. Nucleic Acids Res. 2021, 49, 11855–11867.
- Thomas, E.N.; Kim, K.Q.; McHugh, E.P.; Marcinkiewicz, T.; Zaher, H.S. Alkylative damage of mRNA leads to ribosome stalling and rescue by trans translation in bacteria. eLife 2020, 9, e61984.
- Orita, I.; Futatsuishi, R.; Adachi, K.; Ohira, T.; Kaneko, A.; Minowa, K.; Suzuki, M.; Tamura, T.; Nakamura, S.; Imanaka, T.; et al. Random mutagenesis of a hyperthermophilic archaeon identified tRNA modifications associated with cellular hyperthermotolerance. Nucleic Acids Res. 2019, 47, 1964–1976.
- Alriquet, M.; Calloni, G.; Martinez-Limon, A.; Delli Ponti, R.; Hanspach, G.; Hengesbach, M.; Tartaglia, G.G.; Vabulas, R.M. The protective role of m1A during stress-induced granulation. J. Mol. Cell Biol. 2020, 12, 870–880.
- Tsao, N.; Brickner, J.R.; Rodell, R.; Ganguly, A.; Wood, M.; Oyeniran, C.; Ahmad, T.; Sun, H.; Bacolla, A.; Zhang, L.; et al. Aberrant RNA methylation triggers recruitment of an alkylation repair complex. Mol. Cell 2021, 81, 4228–4242.
- Clark, K.D.; Lee, C.; Gillette, R.; Sweedler, J.V. Characterization of neuronal RNA modifications during non-associative learning in Aplysia reveals key roles for tRNAs in behavioral sensitization. ACS Cent. Sci. 2021, 7, 1183–1190.
- Yang, W.; Meng, J.; Liu, J.; Ding, B.; Tan, T.; Wei, Q.; Yu, Y. The N1-methyladenosine methylome of petunia messenger RNA. Plant. Physiol. 2020, 183, 1710–1724.
- Kuang, W.; Jin, H.; Yang, F.; Chen, X.; Liu, J.; Li, T.; Chang, Y.; Liu, M.; Xu, Z.; Huo, C.; et al. ALKBH3-dependent m1A demethylation of Aurora A mRNA inhibits ciliogenesis. Cell Discov. 2022, 8, 25.
- Ueda, Y.; Ooshio, I.; Fusamae, Y.; Kitae, K.; Kawaguchi, M.; Jingushi, K.; Hase, H.; Harada, K.; Hirata, K.; Tsujikawa, K. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 2017, 7, 42271.
- Woo, H.H.; Chambers, S.K. Human ALKBH3-induced m1A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim. Biophys. Acta. Gene Regul. Mech. 2019, 1862, 35–46.
- Chen, Z.; Qi, M.; Shen, B.; Luo, G.; Wu, Y.; Li, J.; Lu, Z.; Zheng, Z.; Dai, Q.; Wang, H. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019, 47, 2533–2545.
- Esteve-Puig, R.; Climent, F.; Pieyro, D.; Domingo-Domenech, E.; Davalos, V.; Encuentra, M.; Rea, A.; Espejo-Herrera, N.; Soler, M.; Lopez, M.; et al. Epigenetic loss of m1A RNA demethylase ALKBH3 in Hodgkin Lymphoma targets collagen conferring poor clinical outcome. Blood 2020, 137, 994–999.
- Wang, Y.; Wang, J.; Li, X.; Xiong, X.; Wang, J.; Zhou, Z.; Zhu, X.; Gu, Y.; Dominissini, D.; He, L.; et al. N1-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 2021, 12, 6314.
More
