Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Stephan Joel Reshkin and Version 4 by Sirius Huang.
Gemcitabine is still the standard-of-care chemotherapeutic drug for pancreatic ductal adenocarcinoma (PDAC). However, the response rate is quite low. There are multiple mechanisms and participants in gemcitabine resistance.
- resistance to treatment
- gemcitabine
- desmoplastic reaction
- hydroxyurea
- proteasome inhibitors
1. Gemcitabine
Gemcitabine was introduced in pancreatic cancer treatment in 1997 after Burris et al. published their report [1][214]. This was a randomized clinical trial of 126 patients with advanced pancreatic cancer. They found that gemcitabine achieved better results than fluorouracil (5-FU) regarding a modest overall survival improvement and pain control. The mean survival only improved by one month (5.65 for gemcitabine vs. 4.41 for 5FU), but improvements in pain and the Karnofsky index were significant (23.8% for gemcitabine vs. 4.8% for 5FU).
Gemcitabine is still the standard-of-care chemotherapeutic drug for pancreatic ductal adenocarcinoma (PDAC) [2][3][215,216]. However, the response rate is quite low (around 30%), and even lower in advanced cases [4][5][217,218].
It improves average survival by two to three months [6][219], a really poor result. Chemoresistance develops rapidly [7][220] and is therefore the main limiting factor of the drug.
Gemcitabine is used as monotherapy or in combination with other chemotherapeutic drugs [8][221]. Results in combinatorial treatments are slightly better than monotherapy, however the high toxicity involved in combinatorial schemes led to its being used alone in many cases.
2. Mechanisms of Resistance to Gemcitabine
There are multiple mechanisms and participants in gemcitabine resistance. The following factors have been identified:
1. Decrease in deoxycytidine kinase expression or activity [9], thus impeding gemcitabine activation in this rate limiting step [10][11];
-
Decrease in deoxycytidine kinase expression or activity [264], thus impeding gemcitabine activation in this rate limiting step (Figure 4) [265,266];
-
Increased expression of Ribonucleotide Reductase isoform M1 [267,268,269], leading to increased production of nucleotides for DNA synthesis;
-
c-Met activation: inhibition of c-Met with cabozantinib has overcome gemcitabine resistance and increased its cytotoxicity [275,276,277,278,279];
-
CAF-released exosomal miRNA 106-b [284]: CAFs are intrinsically resistant to gemcitabine and transmit this resistance through exosomes containing miRNA 106-b to cancer cells where they target TP53; For a review on miRNAs in pancreatic cancer, read Slotowinski et al. [285].
-
CAF production ofthe chemokine stromal cell-derived factor 1 (SDF1) is able to activate special AT-rich sequence-binding protein 1 (SATBP1), which intervenes in tumor progression and resistance to gemcitabine [286], as shown in the lower panel of Figure 8;
-
SDF-1α produced by stellate cells and secreted in the stroma has the ability to bind the CXCR4 over-expressed in pancreatic cancer cells, activating a pathway that increases survival, reduces apoptosis, and increases expression of IL-6 [287], as shown in Figure 8.
-
Epithelial–mesenchymal transition [290], as the relationship between EMT and gemcitabine resistance is very complex:
-
Gemcitabine-resistant cells acquire EMT phenotype with cancer stem cell characteristics. Notch-2 and Jagged-1 are highly upregulated in these cells [291];
-
Gemcitabine resistance-mediated EMT is in part induced by hypoxia because when HIF-1α is blocked, there is partial reversal of EMT [292];
-
Targeting EMTcan overcome resistance [
-
- Gemcitabine-resistant cells acquire EMT phenotype with cancer stem cell characteristics. Notch-2 and Jagged-1 are highly upregulated in these cells [36];
- Gemcitabine resistance-mediated EMT is in part induced by hypoxia because when HIF-1α is blocked, there is partial reversal of EMT [37];
- miR 233 is a contributing factor to gemcitabine resistance-dependent EMT [38];
- Cells that survive after gemcitabine treatment show increased stemness and EMT markers [39];
- Gemcitabine-induced EMT sustains chemoresistance [40];
- Targeting EMTcan overcome resistance [41].
- ].
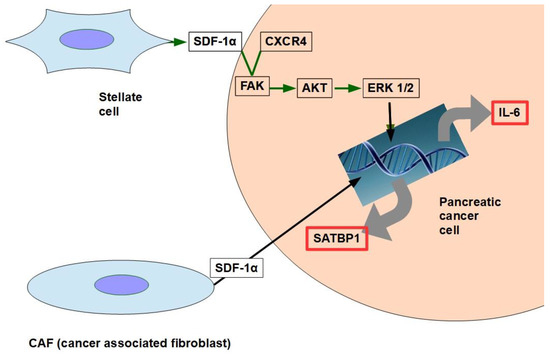
Figure 18. The two pathways shown in the figure have been found to decrease gemcitabine’s cytotoxicity and apoptosis. SDF-1α expression is induced by galectin 1.
Therefore, based on the above evidence, a circuit like the one shown below may represent the chain of events:
Gemcitabine→ EMT→ Resistance to Gemcitabine→ further EMT
- 16.
- 17.
- 18.
-
miRNA 320c through SMARCC1 (SMARCC1 is a protein that forms part of the SWI/SNF complex) [48][303]: This miRNA exerts contradictory actions because it has anti-tumor effects in bladder cancer by downregulating CDK6 [49][304] and in glioma, where it decreases growth and metastasis [50][305]. It was found to decrease canonical Wnt signaling in joints [51][306]. Therefore, miRNAwe 320c can becan considered miRNA 320can anti-oncogenic miRNA [52][307] which, however, promotes gemcitabine resistance.
- 19.
-
miRNA 21 and 221 [53][54][308,309]: miRNA 21 binds to the 3′-UTR region of the Bcl-2 gene, leading to its over-expression and thus inhibiting apoptosis of pancreatic cancer cells [55][310]; antisense miRNAs 21 and 221 restored gemcitabine sensitivity and induced cell-cycle arrest and apoptosis [53][308], while miRNA 200 [56][57][311,312] seems to antagonize miRNA21. miRNA 221 is considered a reliable circulating miRNA for diagnostic purposes [58][313];
- 20.
- 21.
- 22.
- 23.
- 24.
-
miRNA 210 [65][320] downregulates Homeobox protein Hox-A9, which increases NF-kB activity and decreases sensitivity to gemcitabine. However, the role of this microRNA is controversial. Amponsah et al. [66][321] identified miR-210 as a direct suppressor of the multidrug efflux transporter ABCC5; miR210 probably has dualpro- and anti-tumoral effects according to the balance with the oncomucin MUC4. They mutually regulate each other [67][322];
- 25.
The effects of some miRNAs regarding gemcitabine resistance are still a matter of debate. This is the case of miR-421, which seems to be pro-tumoral, reducing the expression of DPC4/SMAD4 [70][325] and at the same time increasing gemcitabine efficacy through decreased SPINK1 expression [71][326]. Furthermore, there is an oleic acid derivative, K73-3, that is able to upregulate miRNA 421 in vitro and in vivo, improving gemcitabine cytotoxicity [72][327]. Therefore, miRNA 421 should be considered an anti-oncogene agent.
In addition to the miRNAs discussed above, there are others without fully proven inhibitor effects on gemcitabine:
- 26.
-
MUC1 and MUC4 [73][74][328,329] (Figure 29): Oncomucins play an important role in gemcitabine resistance that is discussed below. Mucins form a protective envelope surrounding cancer cells and participate in chemoresistance by impeding drug access to the malignant cells. Their production is usually highly increased in pancreatic cancer. There are two mechanisms involved in oncomucin-induced gemcitabine resistance:
-
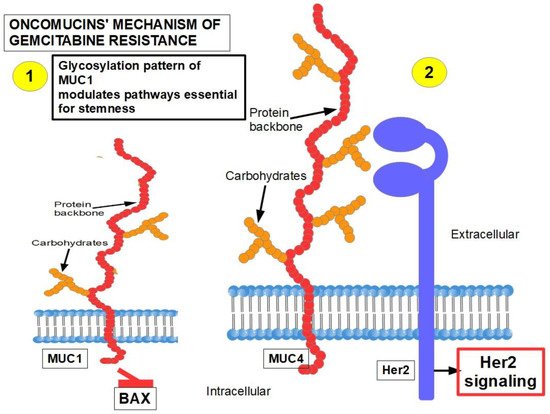
- (1)
-
Direct, by MUC1 inhibiting the apoptotic BAX protein and increasing stemness;
-
- (2)
-
Indirect, by inducing Her 2 signaling.
-
-
-
Tumor-associated oncomucins have a different glycosylation pattern. MUC1 is less glycosylated than MUC4. MUC1-C, the intracellular portion of MUC1, is a driver for the upregulation of PD-L1. Although this immunoescape was found in triple-negative breast cancer, itwe can be hypothesized that pancreatic cancer’s refractoriness to immune-checkpoint inhibitors may be related to MUC1-C. MUC1-C expression also protects the malignant cells against genotoxic attacks in general;
MUC5AC, a facilitator of migration and invasion, also participates in drug resistance by inhibiting TRAIL death pathways.
- 27.
- 28.
- 29.
- 30.
-
ROCK2 (Rho associated protein kinase 2) activity is a cause of acquired gemcitabine resistance [92][347]. The Rhoa/ROCK2 axis promotes migration and metastasis. A pathway has been found in PDAC that shows the long non-coding RNA ZFAS1 inducing metastasis through the Rhoa/ROCK2 axis [93][348]. ZFAS1 is usually overexpressed in PDAC.ROCK inhibitors sensitize pancreatic CSCs to gemcitabine [94][349] and also reduce metastasis;
- 31.
-
Constitutive activation of NF-kB [95][96][350,351]: IL-1α expression is induced by NF-κB, which in turn increases NF-kB in a positive feedback loop, leading to permanent NF-kB activity [97][98][99][100][352,353,354,355]. In addition to the classic PI3K/AKT/NF-kB pathway that is fully operative in PDAC, several other pathways that induce gemcitabine resistance through NF-kB activity have been identified [101][356];
Pancreatic tumors show low miRNA 146-5p expression, impeding regulation of the TRAF6′s 3 UTR segment, thus allowing the pathway shown above. (TRAF6 is the tumor necrosis factor receptor-associated factor 6 that works as an adaptor protein allowing protein–protein interactions) [102][357].
PARP 14 (Poly ADP-ribose polymerase) is highly expressed in PDAC and is associated with poor prognosis. Silencing PARP 14 reduced resistance to gemcitabine.
Clusterin is a protein associated with chemoresistance to different chemotherapeutics. It was found to be increased in PDAC [103][358].
In summary, independently of which pathway activates NF-kB, this transcription factor has the ability to eliminate the pro-apoptotic effects of gemcitabine. Blocking NF-kB can, to a certain degree, decrease gemcitabine resistance [104][359].
- 32.
- 33.
- 34.
- 35.
-
Decreased glutathione peroxidase 1 induced resistance to gemcitabine: Glutathione peroxidase 1 modulates the AKT/GSK3β/Snail signaling axis in PDAC [110][365]. Interestingly, gemcitabine is able to induce the expression of glutathione pathway-related genes which are suspected of generating resistance [111][366];
- 36.
- 37.
- 38.
- 39.
- 40.
-
Activation of Notch signaling [36][117][118][291,372,373] increases therapeutic resistance: This is related to the acquisition of an epithelial-mesenchymal phenotype (see paragraph 15); downregulation of Notch signaling has a chemosensitizing effect [119][374]; Notch-induced chemoresistance to gemcitabine is partly the result of Notch’s ability to alter the intrinsic apoptotic pathway [120][375];
- 41.
-
Hedgehog signaling [121][376]: chemotherapy activates the Hedgehog pathway [24][279], and this activation in turn leads to the expression of stem cell markers such as CD44, SOX2, OCT4, Nanog, and drug efflux proteins of the ATP-binding cassette family. Thus, Hedgehog increases stemness and induces a multidrug resistance phenotype [122][377];
- 42.
-
Cytosolic 5′-nucleotidase 1A over-expression [123][378]: This enzyme is able to reduce gemcitabine’s intracellular metabolites [124][379]. The histone deacetylase inhibitor trichostatin A has been found to synergize with gemcitabine, increasing its cytotoxicity, and importantly, inhibiting 5′-nucleotidase [125][380];
- 43.
-
Pancreatic cancer stem cells [126][381]: Stemness is a key factor in therapeutic failure in most tumors. CSCs do not respond to chemotherapy and are able to replicate the tumor after cytotoxic destruction of sensitive cells. The activation of pancreatic cancer stem cells has shown abilities to promote resistance to gemcitabine. Many of the activators are also involved in resistance. (Figure 310);
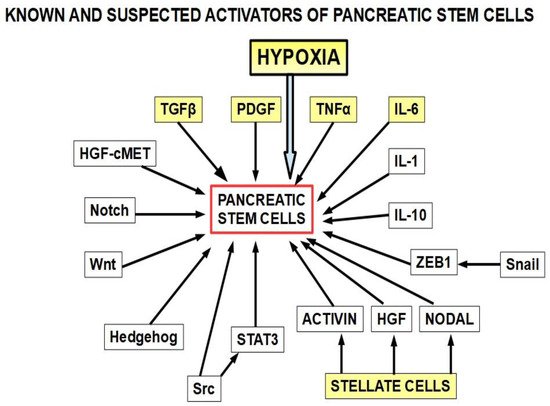 Figure 310. The yellow squares are the known activators of pancreatic cancer stem cells. The other activators (white squares) have also been found to play a role. This diagram is based on references [127][128][129][130][131][132][133][134][135][136][137][138][139][140][141][142][143][144][145][146][382,383,384,385,386,387,388,389,390,391,392,393,394,395,396,397,398,399,400,401]. Importantly, many of the stemness activators are also involved in epithelial–mesenchymal transition.
Figure 310. The yellow squares are the known activators of pancreatic cancer stem cells. The other activators (white squares) have also been found to play a role. This diagram is based on references [127][128][129][130][131][132][133][134][135][136][137][138][139][140][141][142][143][144][145][146][382,383,384,385,386,387,388,389,390,391,392,393,394,395,396,397,398,399,400,401]. Importantly, many of the stemness activators are also involved in epithelial–mesenchymal transition.
- 44.
- 45.
-
Calcyclin-binding protein or Siah-1-interacting protein (CacyBP/SIP) was found to be overexpressed in MDR after gemcitabine treatment. This protein induced P-gp and BCL2 expression reducing apoptosis [149][404]. In addition, CacyBP/SIP knockdown suppresses proliferation in pancreatic cancer by downregulating cyclin E and CDK2 and upregulating Rb and p27 [150][405];
- 46.
- 47.
- 48.
-
The extracellular matrix composition: Laminin and collagen type IV-ECM (mimicking an early tumor ECM) protects from drug-induced apoptosis compared to a collagen I-rich late-tumor ECM;
- 49.
- 50.
-
Autophagy has been shown to be upregulated in PDAC and it plays an important role in resistance to chemotherapy [158][159][413,414]. Autophagy is an inducer of gemcitabine resistance and is probably one of the mechanisms that cells use to survive cytotoxic drugs. Gemcitabine’s cytotoxicity is increased when an autophagy-inhibitor is used simultaneously [160][415]. Pancreatic adenocarcinoma is a very hypoxic tumor, and hypoxia can induce autophagy. Additionally, the expression of high-mobility group box 1 (HMGB1) is an autophagy inducer. Interestingly, gemcitabine upregulates this protein, thus indirectly increasing autophagy [161][416]. In a preoperative setting, when combining the autophagy inhibitor hydroxychloroquine with gemcitabine, 61% of patients showed CA19.9 marker decrease, improved postoperative, and disease-free survival. These findings were particularly evident in the patients with high levels of the autophagy marker LC3-II [162][417]. By blocking autophagy, gemcitabine’s cytotoxic effects were increased and stem cell activity reduced [163][418]. Zeh et al. [164][419] studied two cohorts of preoperative patients, one receiving nab-placlitaxel, and another group with the same medication plus hydroxychloroquine. They found that the resected pancreas in the hydroxychloroquine group had a greater pathologic response and higher immune activity. However, overall survival and disease-free survival was similar in both cohorts. SNHG14 (small nuclear RNA host gene 14) oncogene expression generates a long non-coding RNA that induces autophagy and resistance to gemcitabine [165][420]. This LNC-RNA seems to act as an anti-sense against MiRs involved in anti-tumoral activity;
The conclusion is that there is evidence supporting better results with longer overall survival and disease-free survival by adding autophagy inhibitors to gemcitabine in the resectable cases [166][421]. Evidence in this regard is lacking for inoperable patients.
- 51.
-
Pancreatic tumor microbiota: There is a clinically important population of bacteria and fungi within the pancreas and biliary tree in patients with PDAC and this population is different from the microbiota found in the normal pancreas [167][168][169][422,423,424]. The bacteria present in PDAC show some specificity [170][425]. Regarding gemcitabine, it was found that intratumoral Gammaproteobacteriahad a role in resistance [171][426]. Patients that had some surgical or endoscopic procedure on the pancreas and the biliary tree were prone to host pro-resistance bacteria in the pancreas [172][427], and 5-FU resistance was associated with the presence of Fusobacterium nucleatum in colorectal cancer [173][428]. Fusobacterium is very abundant in PDAC, so it can be hypothesized that it also plays a role in pancreatic chemoresistance. Furthermore, Fusobacterium induces autophagy as part of its chemoresistance mechanism, another frequent finding in PDAC. Fungi have also been found to be a possible cause of gemcitabine resistance [174][429];
- 52.
-
Hypoxia is a key factor in the PDAC phenotype, including proliferation, autophagy, progression, metastasis, as well as resistance to treatment in general, and to gemcitabine in particular [175][430]. The evidence is compelling [176][177][178][179][180][181][182][183][431,432,433,434,435,436,437,438]. A simple example shows the importance of this issue. Hypoxia is expressed through the hypoxia-inducible factors (transcription factors that modulate over 150 genes). Downregulation of HIF-1α with a newly developed molecule, LW6, inhibited autophagic flux, improved the efficacy of gemcitabine, stopped proliferation, and induced cell death [184][439]. LW6 is a novel HIF-1 inhibitor that decreases HIF-1α protein expression [185][186][187][440,441,442];
Hypoxia not only increases resistance to gemcitabine, it also increases gemcitabine-induced stemness [188][443]. Luo et al. [189][444] showed that hypoxia- induced miRNA 301a which in turn promoted gemcitabine resistance through downregulation of T53, thereby integrating hypoxia, miR, and gemcitabine resistance into one pathway. Figure 11.

Figure 411.
Mechanism of hypoxia-induced gemcitabine resistance.
- 53.
-
Increased expression of cytoplasmic ribonucleotide reductase subunit M1 (RRM1) [190][445]: RR is a multimeric enzyme essential for maintaining a high pool of deoxynucleotides for DNA elongation and also for DNA repair. Gemcitabine-resistant pancreatic cancer cells treated with RRM1 inhibitors showed considerable decrease in resistance [13][268]. Patients with high RRM1 levels showed a poorer overall survival with gemcitabine treatment compared with low RRM1- expressing patients [191][446];
- 54.
Figure 512 presents a summary of mechanisms involved in gemcitabine resistance.
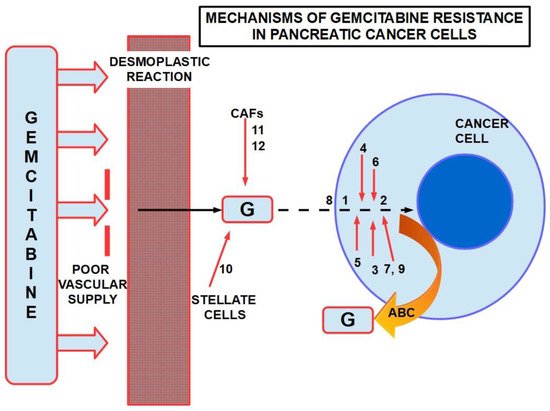
Figure 512. Some mechanisms of resistance to gemcitabine in PDAC. ABC: ATP binding cassette. Poor vascular supply and the desmoplastic reactionare mainly physical barriers. The numbers are chemicals and pathways activated for the escape. ABC re-exports the cytotoxic substances.
