Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Jameel Al-Khayri and Version 2 by Camila Xu.
Fruit trees provide essential nutrients to humans by contributing to major agricultural outputs and economic growth globally. However, major constraints to sustainable agricultural productivity are the uncontrolled proliferation of the population, and biotic and abiotic stresses. Tree mutation breeding has been substantially improved using different physical and chemical mutagens.
- mutagenesis
- TILLING
- genome editing
- targeted mutation
- CRISPR-Cas
- transgene-free
- fruit trees
- biotechnology
1. Introduction
Conventional breeding has been the sole source of genetic improvement in fruit crops for various traits. Classical approaches to introduce a promising trait in an elite cultivar require the introgression of related alleles through multiple generations of selection. For example, the introduction of a disease-resistance trait into a high yielding cultivar is commenced by crossing it with a disease-resistant cultivar, followed by recurrent backcrossing with the elite parent to sustain the genetic potential of the elite cultivar besides conserving the newly introduced resistance allele. Usually, the whole process encompasses several generations to stabilize the resistance allele in the elite background. Fruit crop breeding have certain limitations, which may include outcross reproduction, prolonged juvenility, and enormous genome landscapes [1][2]; therefore, it requires decades to improve such traits. The obligate outcrossing nature of fruit trees amalgamates classical breeding for genotypic and phenotypic traits. A relevant example to illustrate this dilemma is the development of resistance to apple scab. Hough et al. [3] conducted a wide range of crosses between an elite apple cultivar and a genetically compatible wild-type cultivar as the source of resistance to apple scab. However, over several decades of continuous breeding, the resultant cultivars lost the fruit quality traits [4]. The application of marker-assisted selection, such as marker-assisted breeding (MAB), marker-assisted selection (MAS) and genome-wide association mapping (GWAS) for quantitative trait loci (QTLs), may contribute to shorten the selection process, but not bypass the generations of backcrossing [5]. For example, apples, cucumbers, mandarins, peaches, and strawberries have been substantially improved [6][7][8]. Fast-track breeding approaches may possibly overcome extreme juvenility in fruit trees via the transgenic expression of the desired genes. The breeding time for fruit trees can be shortened to one-fifth of the conventional crossbreeding approaches [9]. For example, the apple cultivar ‘Pinova’ was transformed to impart early flowering by expressing a MADS-box gene from Betula pendula [10]. In another study, null segregants of fire blight and apple scab resistant apples were generated within seven years [11]. Similarly, Endo et al. [7] successfully substituted the genetic background of mandarin through an integrated transgenic approach and MAS to transfer CTV resistance from a transgenic trifoliate orange. However, to obtain the null segregants, the transgene should be segregated out from the elite parental background through backcrossing with the recurrent parent. The detachability of the T-DNA transgene can be confirmed through comparative genomic hybridization (CGG) and next-generation sequencing (NGS) approaches [9]. In fast-track breeding, MAS plays a critical role to increase the selection efficiency in the backcrossed progenies.
Several new strategies were introduced in the middle of the 20th century to enrich the genetic diversity of fruit trees. Mutagenesis has been used to facilitate plant breeding since the 1920s with the discovery that mutations induced with physical (gamma irradiation) or chemical mutagen treatment can be inherited [12]. Importantly, with the discovery of X-rays, a subsequent series of induced mutations were conceptualized in plants and animals (Figure 1). In 1934, the first commercial mutant tobacco variety was produced [13]; since then, mutant crop cultivars have been continuously registered globally (Figure 2A,B). It was not until 1963 that the first mutated apple cultivar “Mori-hou-fu 3A” was developed in Japan through gamma rays. The following year, a sweet cherry (Prunus avium L.) cultivar “Compact Lambert” was developed in Canada (Table 1). The use of chemical mutagen “EMS” was successfully applied for mutating apples to develop another mutant cultivar “Belrene” in France in 1970. The application of physical mutagens was also successful in tree breeding. For example, grapefruit (Citrus × paradisi) cultivar “Rio Red” and clementia (Citrus celementina) cultivar “Nero” were developed using thermal neutrons and fast neutrons in 1970 and 2006 in USA and Spain, respectively. Various hybridization methods were also developed to produce hybrids between sexually incompatible species by disrupting the meiotic cell division to form polyploids, followed by the restoration of meiosis. Additionally, hybridization approaches also included chromosomal additions/subtractions or the fusion of protoplasts from sexually incompatible species [14]. The genetic background of the elite crop cultivars was further broadened through chemical or physical mutagenesis to increase the desirable alleles in the elite lines.
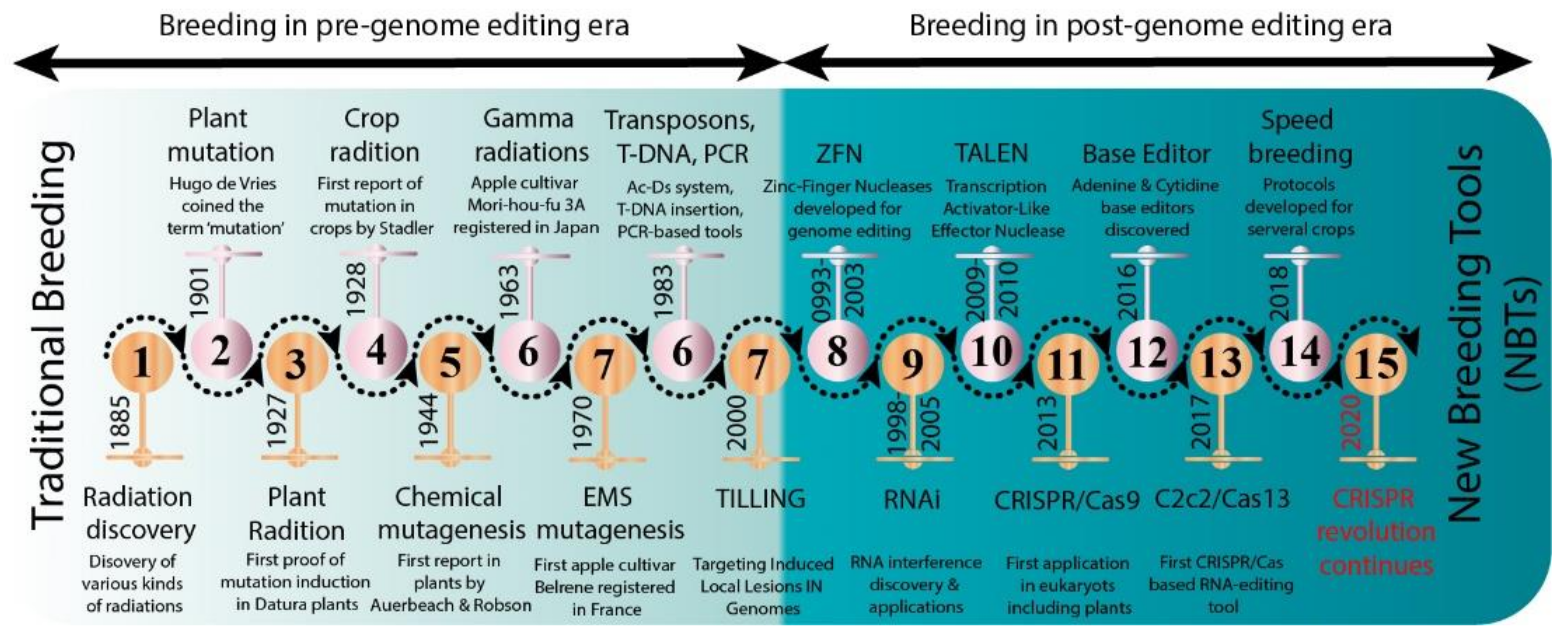
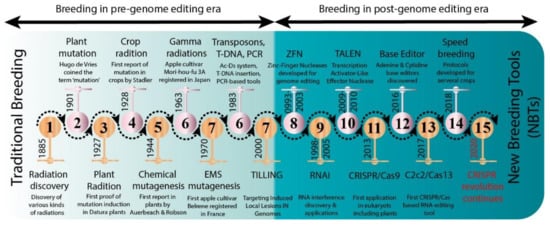
Figure 1. Historic timeline for mutagenesis in plants.
Historic timeline for mutagenesis in plants.
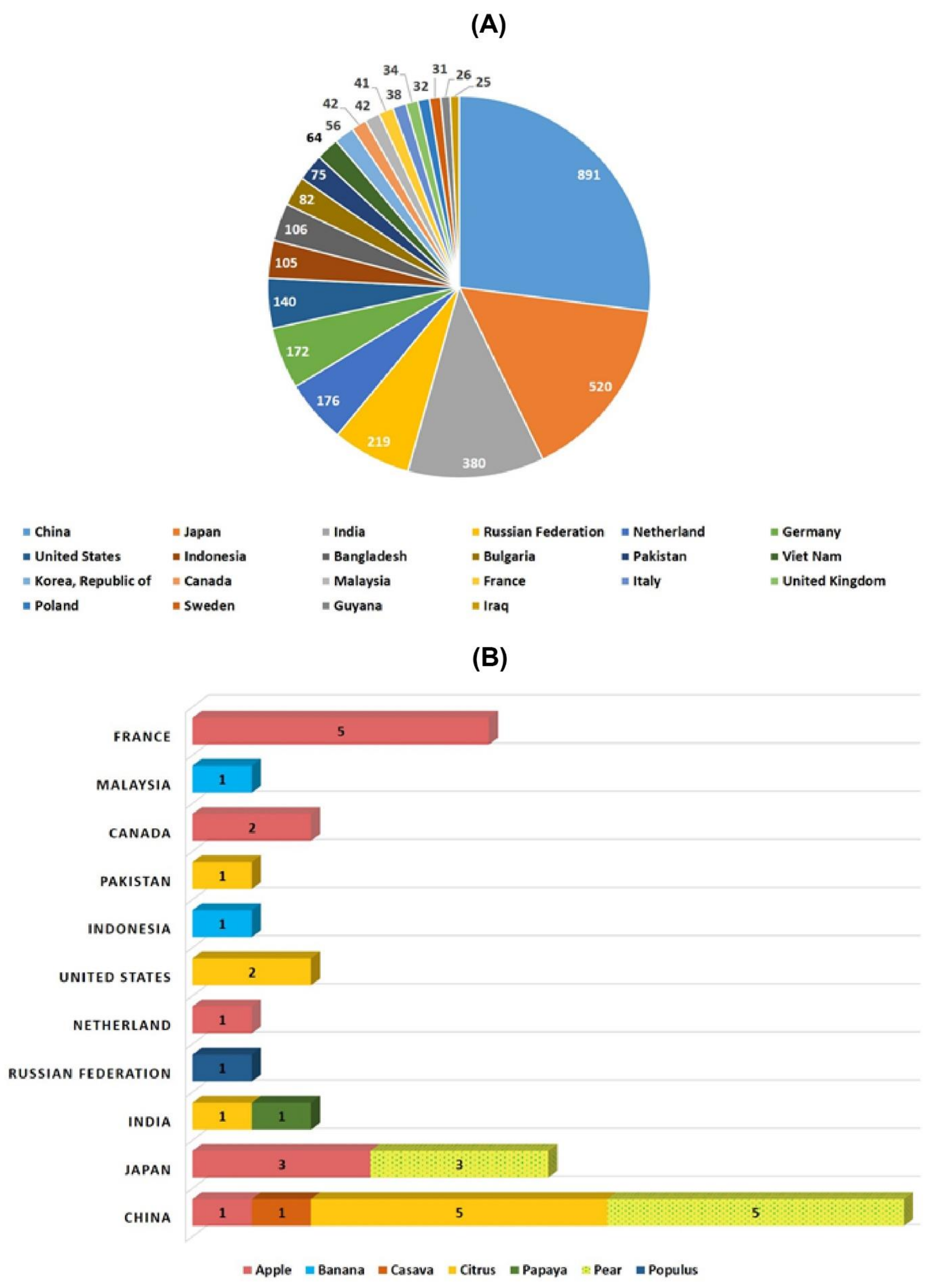
Figure 2. Number of mutant varieties released in top 22 countries (A) and number of mutant tree varieties of assorted fruit species in selected countries (B).
Table 1.
Country-wise varietal approval developed by mutagen treatment.
| Common Name | Botanical Name | Variety | Mutagen Source | Country | Year of Registration | |
|---|---|---|---|---|---|---|
| Almond | Prunus dulcis | Mill. | Supernova | Gamma rays (30 Gy) | Italy | 1987 |
| Apple | Malus pumila | Mill. | Mori-hou-fu 3A | Gamma rays (30 Gy) | Japan | 1963 |
| Senbatsu-Fuji-2-Kei | Gamma rays (60 Gy) | 1985 | ||||
| Belrene | EMS | France | 1970 | |||
| Blackjoin BA 2 520 | Gamma rays (50 Gy) | 1970 | ||||
| Courtagold | 1972 | |||||
| Courtavel | 1972 | |||||
| Lysgolden | 1972 | |||||
| Donghenghongpingguo | Gamma rays (250 Gy) | China | 1987 | |||
| Dovar | X-rays (30–35 Gy) | Netherlands | 1978 | |||
| Golden Haidegg | Gamma rays (50 Gy) | Austria | 1986 | |||
| James Grieve Double Red | Gamma rays (62 Gy) | Czech Republic | 1995 | |||
| McIntosh 8F-2-32 | Gamma rays | Canada | 1970 | |||
| Shamrock | Gamma rays | Canada | 1986 | |||
| Apricot | Prunus armeniaca | L. | Early Blenheim | thermal neutrons (thN) | Canada | 1970 |
| Banana | Musa paradisiaca | L. | Klue Hom Thong KU1 | Gamma rays (25 Gy) | Thailand | 1985 |
| Novaria | Gamma rays (60 Gy) | Malaysia | 1995 | |||
| AL-BEELY | Gamma rays | Sudan | 2007 | |||
| Pirama 1 | Gamma rays (30 Gy) | Indonesia | 2019 | |||
| Fuxuan 01 | Gamma rays | China | 2005 | |||
| Clementina | Citrus celementina | L. | Nero | Fast neutron (6 Gy) | Spain | 2006 |
| Neufina | 2010 | |||||
| CLEMENVERD | Fast neutron (5 Gy) | Spain | ||||
| Ficus | Ficus benjamina | L. | Golden King | X-rays (25 Gy) AND Gamma rays (20 Gy) | Belgium | 1980 |
| Golden Princess | ||||||
| Fig | Ficus carica | L. | Bol (Abundant) | Gamma rays (50–70 Gy) | Russian Federation | 1979 |
| Grapefruit | Citrus paradisi | Macf. | Rio Red | Thermal neutrons (thN) | United States | 1970 |
| Star Ruby | 1984 | |||||
| Indian Jujube | Ziziphus mauritiana | Lamk. | Dao tien | MNH (0.02–0.04%) | Viet Nam | 1986 |
| Ma hong | ||||||
| Japanese pear | Pyrus pyriforia | Nak. | Gold Nijisseiki | Gamma rays (0.12–0.15 Gy) | Japan | 1991 |
| Kotobuki Shinsui | Gamma rays (80 Gy) | 1997 | ||||
| Osa Gold | 1997 | |||||
| Lemon | Citrus limon | L. | Eureka 22 INTA | X-rays (10 Gy) | Argentina | 1987 |
| Loquat | Eriobotrya japonica | L. | Shiro-mogi | Gamma rays (200 Gy) | Japan | 1982 |
| Mandarin | Citrus reticulata | L. | Zhongyu 7 | Gamma rays (100 Gy) | China | 1985 |
| Zhongyu 8 | 1986 | |||||
| Hongju 420 | ||||||
| NIAB Kinnow | Gamma rays (20 Gy) | Pakistan | 2017 | |||
| PAU Kinnow-1 | Gamma rays (30 Gy) | India | 2017 | |||
| Mulberry | Morus alba | L. | Sangfu 1 | Gamma rays (75 Gy) | China | 1974 |
| Fuzaofeng | Gamma rays (5 Gy) | 1992 | ||||
| Ji 7681 | N2 laser | 1988 | ||||
| Fusang 10 | Gamma rays | 1980 | ||||
| Shansang 871 | Gamma rays (60 Gy) | 1994 | ||||
| Shigu 11-6 | Gamma rays (100 Gy) | 1995 | ||||
| Lala Berry | Colchicine | Japan | 2003 | |||
| Pop Berry | Colchicine | 2004 | ||||
| S54 | EMS | India | 1974 | |||
| Orange | Citrus sinensis | L. | Hongju 418 | Gamma rays (100 Gy) | China | 1983 |
| Xuegan 9-12-1 | ||||||
| Valencia 2 INTA | X-rays (20 Gy) | Argentina | 1987 | |||
| IAC 2014 | Gamma rays (40 Gy) | Brazil | 2016 | |||
| Papaya | Carica papaya | L. | Pusa nanha | Gamma rays (150 Gy) | India | 1987 |
| Peach | Prunus persica | L. | Magnif 135 | Gamma rays | Argentina | 1968 |
| Shaji 1 | CO | 2 | laser | China | 1985 | |
| Shaji 2 | ||||||
| Fuku-ekubo | Gamma rays (30 Gy) | Japan | 1996 | |||
| Shimizu Hakutou RS | 2004 | |||||
| Plovdiv 6 | Gamma rays (10 Gy) | Bulgaria | 1981 | |||
| Pear | Pyrus communis | L. | Fuxiangyanghongdli | Gamma rays (2.5 Gy) | China | 1983 |
| Chaofu 1 | 1989 | |||||
| Chaofu 10 | ||||||
| Chaofu 10 | ||||||
| Chaofu 2 | ||||||
| Plum | Prunus domestica | L. | Spurdente-Ferco | Gamma rays | France | 1988 |
| Pomegranate | Punica granatum | L. | Karabakh | Gamma rays (50–70 Gy) | Russian Federation | 1979 |
| Khyrda | ||||||
| Sour cherry | Prunus cerasus | L. | Plodorodnaya Michurina | X-rays | Russian Federation | 1977 |
| Karlik Samorodka | Gamma rays | 1979 | ||||
| Polukarlik Orlovskoi Rannei | ||||||
| Polukarlik Turgenevki | ||||||
| Nishina Zao (DT2008) | Ion beams | Japan | 2009 | |||
| Sweet cherry | Prunus avium | L. | Compact Lambert | X-rays (40 Gy) | Canada | 1964 |
| Compact Stella 35B-11 | 1974 | |||||
| Van 2D-14-11 | 1972 | |||||
| Lapins | X-rays | 1983 | ||||
| Lambert 2B-17-18-EC | X-rays (50 Gy) | 1972 | ||||
| Stella | 1968 | |||||
| Stella 16A-7 | 1972 | |||||
| Sunburst | 1983 | |||||
| Sumste Samba | Gamma rays | 2000 | ||||
| ALDAMLA | Gamma rays (25 Gy) | Turkey | 2014 | |||
| BURAK | Gamma rays (50 Gy) | |||||
| Burlat C1 | Gamma rays | Italy | 1983 | |||
| Nero II C1 | ||||||
| Ferrovia spur | X-rays (4 Gy) | 1992 | ||||
| Super 6 | Colchicine | Japan | 1997 | |||
| Roman Nishiki | 2002 |
The advent of plant transformation approaches including Agrobacterium-mediated, particle bombardment or electroporation-mediated and chemical transfections through protoplasts allowed the development of transgenic plants with the specific genetic constructs from any biological source. Although these methods involve in vitro culturing, many fruit tree species are recalcitrant to transformation and regeneration. However, fruit crops amenable to in vitro culturing were successfully transformed to directly introgress new genes without recurrent backcrossing [15]. However, the resulting transgenic plants faced regulatory complexities and gave rise to a dichotomy between the product and process regulatory framework [16]. These legalities have some practical implications because even if the gene transfer is intraspecific (involving plants from the same species), it still left some cargo of a transgene such as the remnants of the genetic markers or parts from the bacterial plasmid itself or the T-DNA of A. tumefaciens [17].
Genome editing (GE) technology has revolutionized fruit crop breeding [18]. GE encompasses three types of specific site-directed nucleases (SNDs)—i.e., SDN-1, SDN-2, and SDN-3—to cause double-stranded breaks (DSBs) at pre-defined genome targets. The major advantage of SDNs is the targeted DNA cleavage and the subsequent use of cellular machinery to introduce the desired change during the repairing processes. These DSBs are imprecisely repaired by the cell DNA repair mechanism and cause insertion and deletion (indels) mutation over all dysfunctions of the gene of interest without introducing any foreign element into the genome. The entire process is well-regulated and, at the sequence level, the indel mutants cannot be distinguished from natural variations and/or irradiated or chemical mutants [19]. Four SDN-based GE techniques include meganucleases, zing-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the most recent clustered regularly interspaced short palindromic repeat/CRISPR-associated proteins (CRISPR/Cas), respectively. These GE techniques are based upon biological molecules with a DNA-binding domain and the cleavage activity (through nucleases). The mutants developed with chemical or physical mutagen treatment are usually hemizygous and their homozygosity is achieved through either filial segregations or recurrent backcrosses to fix the introduced mutations. Nevertheless, this is not a case with site-directed or targeted mutagenesis using GE, and GE shared another edge that is its multiplexing capabilities—i.e., simultaneous targeting of multiple genes or copies of a gene. This characteristic can be extremely useful to target homologous genes in polyploid fruit trees. The recent advancements in GE to substitute a single nucleotide allows individual base swapping in a DNA strand [20]. These developments in GE can help to overcome the GMO regulatory frameworks because it leaves no trace of a transgene or exogenous source in the targeted genome. Moreover, there is evidence that the remnants of Agrobacterium T-DNA have a role in the evolution of some plant families [21]. Thus, the boundary between natural and engineered crop species may become more blurred after such evidence and ultimately gain the attention of the scientific community to revise the regulatory framework, at least for GE crops.
2. Tree Breeding under Climate Change
The response of trees to any global climate change scenario is a pressing question for natural vegetation and man-made plantations [22]. Climate change is a major threat to tree plantations due to fluctuations in annual precipitation, drought, heat, salinity, and enhanced insect infestations [23]. Investigations to explore the ability and genetic basis of adaptation to global climate change in ecologically and industrially distinct tree species to cope with abiotic and biotic stresses are key research lines in plant science [24]. However, this knowledge has rarely been translated into conserving the genetic resources or bringing the genetic improvements to woody perennials.
The objective of most tree breeding programs is to gradually improve tree populations through recurrent selection cycles and verifications [25]. Traditionally, tree breeding mostly relies on phenotypically selecting superior candidates from the natural or planted stands. It constitutes the base population and further selection builds a pool of selected population with elite donors. Furthermore, these selected populations are then tested for progeny trials and the reselection of parents [26]. However, such selections may cause genetic erosions in the overall populations due to inbreeding depression. The production of hybrids and subsequent backcrossing may accelerate classical breeding with the aim of harnessing heterotic effects by virtue of dominance or over-dominance, tree adaptability and increased yield [27]. Among other potential applications, hybrid breeding has been widely applied to maximize the tree crown perimeter, tree height, conferring resistance to Fusarium spp. [28], and to chestnut blight from wild donor tree plants into American chestnut populations [29].
The most promising alternative approach towards tree breeding is molecular- marker-assisted selection (MAS) and molecular-assisted breeding (MAB) [30]. MAS and MAB tools can be effective in pyramiding simple Mendelian traits regulated by a few genes but have limited utility for selection against complex genetic traits in trees [31]. Moreover, MAS and MAB cannot be effective due to fluctuations in allelic frequencies over generations and therefore cannot explain genetic variations for complex traits [1]. To circumvent these limitations, the use of the genomic selection (GD) approach is suitable rather than phenotypic selection-based traditional breeding using MAS and MAB. Despite having a relatively short history, the technique has been successfully implemented in plant breeding. It can substantially reduce the long breeding cycles for tree breeding and positively enhance the genetic gain over time [31]. In the GS approach, a large number of molecular markers are used to analyze the cumulative effects of QTLs evenly distributed over the genome. Therefore, it makes GS much more efficient for tree breeding due to the possibility to assess the individual genomic estimated breeding value (GEBV) of a single plant. It involves four basic steps: (a) phenotyping and genotyping of the selected individuals from a breeding population, (b) generation of genomic prediction models, (c) model validation on the test population, and d) prediction of GEBV for non-phenotyped individuals and further selection [32]. Unlike MAS, there are no pre-requisites in GS for a prior information about marker linkage, or QTL localizations in the genome and their relative phenotypic effects [33].
3. Mutagenesis as a Source of Genetic Variability in Tree Plants
Genetic improvement through conventional breeding necessarily requires recurrent selection cycles in fruit trees [6] (Figure 3). However, a major limitation is the large number of crosses and the development of subsequent filial generations. This is even more challenging in fruit trees such that recurrent selection may take decades of continuous breeding efforts [34]. The lengthy breeding process can be accelerated in fruit frees with more advanced techniques such as MAS and GWAS for QTLs [35]. For example, many quality- and yield-related traits have been improved in apple, banana, mandarin, peach, and strawberry through conventional breeding coupled with mutagenesis, MAS, genetic engineering, MAB and others [6][7][9]. The genetic improvements in fruit trees are, however, progressing at a slower pace, but the availability of pangenomes, broader understanding of genotypic and phenotypic interactions and fast-track breeding may hasten the development of fruit tree cultivars with better genetic makeup.
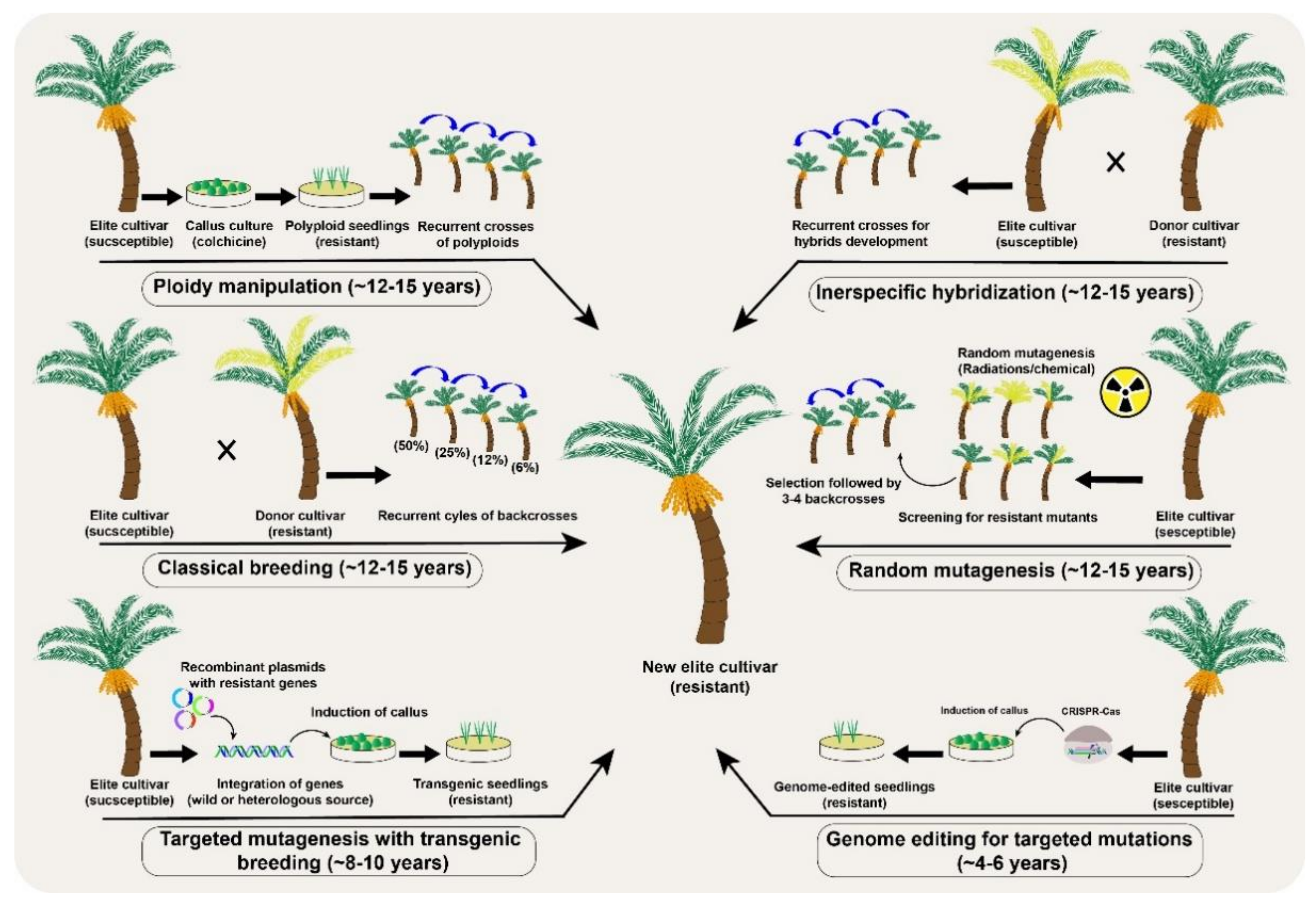
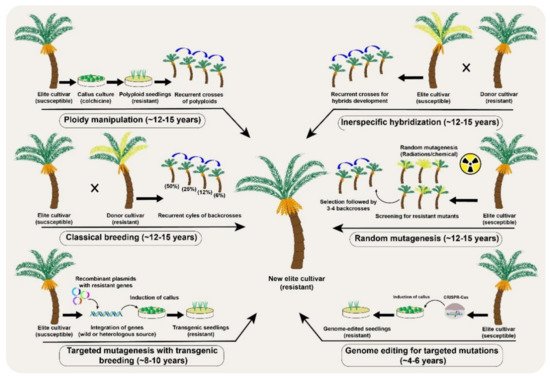
Figure 3. A comparative analysis of different conventional and the new breeding tools (NBTs) to modify desirable genetic modifications in a date palm (Phoenix dactylifera L.) fruit crop.
Genetic improvement through conventional breeding is limited to sexually compatible crop plants [36]. Nevertheless, the genetic diversity of self-incompatible plants can be increased by mutagen treatment (physical or chemical) to induce new mutations in cultured cells, seeds, seedlings, or sometimes whole plants. Random mutations are preferred in seeds because the small number of cells in the developing embryo makes mutagenesis much easier and produces fewer chimeric plants [37]. During somatic mutations, a fewer number of cells or limited sectors in the apical meristem are affected, creating chimeric mutated plants. Such sectorial mutations involve genetic differences in either one or two layers of cells and is more frequent in vegetatively propagated fruit trees [38]. An effective way to dissociate chimerism in vegetatively propagated plant is through organogenesis or embryogenesis [39][40]. The mutation frequency and population structure of the mutants directly depend upon the type of mutagen and the time of exposure. Irrespective of the used mutagens, the ultimate induced mutations are random and therefore require a large screening population to identify the desired mutants [41]. Mutation breeding is advantageous over conventional breeding because it precludes segregation progenies while improving the genetic make-up during selection cycles.
High energy X-ray radiations were the earliest mutagens used to induce desired traits in fruit trees [37]. Currently, X-ray mutagenesis has been either replaced or supplemented with other more advanced physical mutagens such as fast neutrons, ionizing radiations and gamma rays. Besides bringing about beneficial mutations from single-nucleotide replacements to chromosomal aberrations, these mutagens may trigger DNA damage directly or indirectly in the form of oxygen radicals [42]. Physical mutagenesis has been successfully used to improve a number of traits in fruit trees—for example, improved heat tolerance in pineapple [43], self-fertile in cherry fruits, fruit color in apple, bunch size in banana, short-statured papaya plants, disease resistant pear and growth earliness in grapevine [44].
Among the chemical mutagens, ethylmethanesulfonate (EMS) is the most widely employed alkylating agent in fruit crops [45], including banana and peach [46]. However, it is not suitable for vegetatively propagated fruit trees and perennial allogamous fruit trees because of their heterozygous genomes and prolonged life cycle. Although, chemical mutagens are extremely efficient in inducing desirable mutagenesis in whole plants or seeds, it is not recommended for tissue-cultured plants due to their extreme toxicity [37]. Chemical mutagens predominantly cause hemizygous point mutations and successive backcrosses are necessary to obtain a homozygous line and to stabilize the mutated gene of interest [47]. On the other hand, physical mutagenesis has a high risk of a collateral effect on non-targeted genes because the impact of physical mutagens produces multisite mutations of various sizes. For example, the use of fast neutron mutagenesis causes large deletions in the genome besides translocations and chromosomal loss [48][49]. Chemical mutagens are more affordable; however, these carry serious health and environmental risks. Moreover, chemically induced mutations are genetically less stable than physical mutations.
Polyploidy is another interesting natural phenomenon in plant evolution, adaptation, and speciation, which can also be induced using colchicine, for genetic improvements. Colchicine application induces autopolyploidy by blocking mitosis without interfering with DNA replication and ultimately doubles the chromosome numbers (Figure 3). The generation of triploid dessert apple and tetraploid grapevine cultivars are successful examples of autopolyploids in fruit crops [50]. Interspecific hybridizations have also been tested in some citrus cultivars, including the formation of natural hybrids [51]. However, as in conventional breeding, if the hybrids are fertile in perennial fruit trees, multiple backcrosses are still needed to remove the undesirable genetic background of the recessive parent. For example, scab resistance in apple took more than 40 years [52], and the enhancement of sugar and antioxidants levels in elderberries took at least 10 years through the interspecific hybridization approach [53].
Somaclonal variation is a natural phenomenon occurring during in vitro tissue culturing and can produce useful genetic variations in plants [54]. It includes DNA-related genetic or epigenetic variations, which induce phenotypic changes distinguishable from the original parent. Major causes include but are not limited to prolonged in vitro culturing, tissue culturing media composition, the presence of phytohormones and certain other mechanical factors during culturing [55]. Somaclones can be detected through morphological assessments of the off-type regenerants, biochemical response of explants, fingerprinting with protein or isozymes-based markers, and cytogenetic assessment [56][57]. In addition, more advanced DNA- or transposon-based molecular markers [58] and the use of next-generation sequencing (NGS) screening have also been successfully applied to detect somaclonal variations in fruit tree breeding.
TILLING as a Powerful Tool in Mutation Breeding
Numerous significant genes from older mutant cultivars continue to serve as a foundation for modern gene pools in commercial cultivars. Nonetheless, the burden of unwanted genetic mutations and the development of new breeding tools (NBTs) have had an effect on the use of random mutation techniques [59]. Recent advancements in screening methods enable the detection of SNPs and complex traits at the molecular level, which are otherwise difficult to discern with conventional screening methods. The utilization of mutagenesis underwent a huge change with the development of TILLING (targeting induced local lesions in genomes) as a high-throughput mutant screening technique to identify point mutations at a specific locus in the mutated genome [60]. The TILLING technique redirected mutation breeding away from laborious forward genetics approaches to reverse genetics approaches, allowing plant breeders to detect mutations in known genes. Furthermore, TILLING has been accompanied with the more advanced next-generation sequencing (NGS) techniques to provide more practical solutions to bypass extensive mutant screening for the selected genes [61].
The major mutation screening methods in TILLING include celery nuclease (CEL I) [62], high-resolution melting (HRM) [63] and NGS [64]. The mismatch-specific CEL I method is a popular TILLING technique, which is coupled with the LI-COR gel analyzer system. The HRM incorporates the PCR technique in which the monitoring of dsDNA product is monitored with a dsDNA-specific fluorescent dye followed by the formation of a high-resolution melting curve. The more advanced NGS technique has further facilitated the mutant screening in a TILLING population through whole-genome sequencing, de novo assembly and resequencing tools.
The basic procedure of TILLING includes mutation induction through chemical, physical or biological agents to produce an M1 population. These M1 plants are then allowed to self-pollinate and generate M2 plants. Total genomic DNA is isolated and subjected to eightfold DNA pooling followed by PCR amplification of the gene of interest. The recurrent heating and cooling steps form heteroduplexes, which are then incubated with CEL I endonuclease to cleave mismatches in these heteroduplexes. The cleaved DNA products are separated on a denaturing gel electrophoresis and the fluorescence is detected with a LI-COR DNA analyzer. The induced mutations are then verified by sequencing of the polymorphic individuals, respectively [65]. Although the CEL I -based TILLING platform has been widely used, the critical steps such as enzymatic digestions, cloning and gel electrophoresis make it time consuming. Moreover, insufficient genome sequence information of many plant species affects the efficacy of this TILLING platform [66]. Contrarily, the HRM-based TILLING offers more accurate, sensitive, and cost-effective mutant screening through PCR and analysis of the DNA melting curve. Nevertheless, detection of small insertions and deletions is difficult and limited to amplicons with a size <450 bp with HRM [67]. The NGS-based TILLING platform is comparatively a potential screening method with more accurate mutant screening. However, the high cost, the generation of a large sequencing dataset and the requirement of sophisticated bioinformatics tools still pose major challenges to its adoption in studying the genetics and genomics of mutagenic studies [65].
4. Genomics and Genetic Engineering Perspectives of Trees
The genetic improvement of the tree plant genome can be accelerated through two distinct approaches: MAB through quantitative trait loci (QTL) mapping, and direct gene transfer through genetic engineering. The whole genomes of many tree plants have been completely sequenced; consequently, comprehensive genetic architecture of useful genetic traits are now available, which can be helpful for marker-assisted breeding, MAB [68][69]. The availability of such datasets can widely assist in genetic expression, and functional and comparative genomics. Moreover, recent developments in –omics and NGS technologies and, in parallel, more advanced bioinformatics tools, can expedite in-depth molecular studies in trees [70][71]. The transcriptomic, proteomic and metabolomics data sets of woody plants are dynamically bridging the gaps between tree genomes and genetic expression studies.
Genetic transformation can be improvised by inserting single or multiple genes directly into the elite background across the species or genus without long cycles of selections and screening [72][73], e.g., herbicide tolerance in populous [74]. The first application of genetic engineering in fruit trees was in papaya when papaya varieties ‘Sunset’ and ‘Kapoho’ were genetically modified by inserting the capsid protein (CP) gene of papaya ringspot virus (PRSV) to confer viral resistance. Consequently, the first transgenic papaya cultivar was developed in 1998 [75]. Recently, the USA approved a non-browning arctic apple cultivar [76][77]. Several other genetic traits for fruit quality, tree physiology and abiotic stress tolerance have been successfully engineered for transgenic apple, banana, papaya, and pineapple [73][75]. Current transgenic fruit trees approved in the USA include papaya against PRSV, plum against plum pox virus (PPV) [78], apple with the non-browning trait [79] and pineapple cultivar ‘Pinkglow’ [80]. Transgenic papaya plants have been successfully engineered to alter elite traits related to tree growth, nitrogen metabolism, lignin contents and abiotic stress tolerance [81][82]. Moreover, resistance in papaya was also conferred against phytophthora blight, papaya dieback disease (PDBD) and papaya ringspot virus (PRSV) in several studies [83]. Among non-transgenic approaches, dsRNA-mediated protection strategies have also been practiced in papaya against PRSV [84]. Similarly, eucalyptus species have also been genetically transformed to introduce genes from endogenous or heterologous sources to modify their salt tolerance status and secondary cell wall constituents [85]. Many pine softwood tree species have also been utilized for transgenic developments for various traits [69].
References
- Grattapaglia, D.; Silva-Junior, O.B.; Resende, R.T.; Cappa, E.P.; Muller, B.S.F.; Tan, B.Y.; Isik, F.; Ratcliffe, B.; El-Kassaby, Y.A. Quantitative genetics and genomics converge to accelerate forest tree breeding. Front. Plant Sci. 2018, 9, 1693.
- Burdon, R.D.; Klapste, J. Alternative selection methods and explicit or implied economic-worth functions for different traits in tree breeding. Tree Genet. Genom. 2019, 15, 79.
- Hough, L.F.; Shay, J.R.; Dayton, D.F. Apple Scab Resistance from Malus-Floribunda Sieb. Proc. Am. Soc. Hort. Sci. 1953, 62, 341–347.
- Schouten, H.J.; Krens, F.A.; Jacobsen, E. Cisgenic plants are similar to traditionally bred plants—International regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO Rep. 2006, 7, 750–753.
- Semagn, K.; Bjornstad, A.; Ndjiondjop, M.N. Progress and prospects of marker assisted backcrossing as a tool in crop breeding programs. Afr. J. Biotechnol. 2006, 5, 2588–2603.
- Moose, S.P.; Mumm, R.H. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008, 147, 969–977.
- Omura, M.; Shimada, T. Citrus breeding, genetics and genomics in Japan. Breed. Sci. 2016, 66, 3–17.
- Feng, S.J.; Zhang, J.P.; Mu, Z.H.; Wang, Y.J.; Wen, C.L.; Wu, T.; Yu, C.; Li, Z.; Wang, H.S. Recent progress on the molecular breeding of Cucumis sativus L. in China. Theor. Appl. Genet. 2020, 133, 1777–1790.
- Endo, T.; Fujii, H.; Omura, M.; Shimada, T. Fast-track breeding system to introduce CTV resistance of trifoliate orange into citrus germplasm, by integrating early flowering transgenic plants with marker-assisted selection. BMC Plant Biol. 2020, 20, 224.
- Elo, A.; Lemmetyinen, J.; Novak, A.; Keinonen, K.; Porali, I.; Hassinen, M.; Sopanen, T. BpMADS4 has a central role in inflorescence initiation in silver birch (Betula pendula). Physiol. Plant. 2007, 131, 149–158.
- Schlathölter, I.; Jänsch, M.; Flachowsky, H.; Broggini, G.A.L.; Hanke, M.-V.; Patocchi, A. Generation of advanced fire blight-resistant apple (Malus × domestica) selections of the fifth generation within 7 years of applying the early flowering approach. Planta 2018, 247, 1475–1488.
- Stadler, L.J. Genetic effects of x rays in maize. Proc. Natl. Acad. Sci. USA 1928, 14, 69–75.
- Acquaah, G. Principles of Plant Genetics and Breeding; John Wiley & Sons: Hoboken, NJ, USA, 2009.
- Sink, K.; Jain, R.; Chowdhury, J. Somatic cell hybridization. In Distant Hybridization of Crop Plants; Springer: Berlin/Heidelberg, Germany, 1992; pp. 168–198.
- Kohli, A.; Christou, P. Stable transgenes bear fruit. Nat. Biotechnol. 2008, 26, 653–654.
- Ammann, K. Genomic misconception: A fresh look at the biosafety of transgenic and conventional crops. A plea for a process agnostic regulation. New Biotechnol. 2014, 31, 1–17.
- Twyman, R.M.; Kohli, A.; Stoger, E.; Christou, P. Foreign DNA: Integration and expression in transgenic plants. In Genetic Engineering: Principles and Methods; Setlow, J.K., Ed.; Springer: Boston, MA, USA, 2002; pp. 107–136.
- Ghogare, R.; Williamson-Benavides, B.; Ramirez-Torres, F.; Dhingra, A. CRISPR-associated nucleases: The Dawn of a new age of efficient crop improvement. Transgenic Res. 2020, 29, 1–35.
- Zhu, C.; Bortesi, L.; Baysal, C.; Twyman, R.M.; Fischer, R.; Capell, T.; Schillberg, S.; Christou, P. Characteristics of genome editing mutations in cereal crops. Trends Plant Sci. 2017, 22, 38–52.
- Monsur, M.B.; Shao, G.; Lv, Y.; Ahmad, S.; Wei, X.; Hu, P.; Tang, S. Base editing: The ever expanding clustered regularly interspaced short palindromic repeats (CRISPR) tool kit for precise genome editing in plants. Genes 2020, 11, 466.
- Quispe-Huamanquispe, D.G.; Gheysen, G.; Kreuze, J.F. Horizontal gene transfer contributes to plant evolution: The case of Agrobacterium T-DNAs. Front. Plant Sci. 2017, 8, 2015.
- Isabel, N.; Holliday, J.A.; Aitken, S.N. Forest genomics: Advancing climate adaptation, forest health, productivity, and conservation. Evol. Appl. 2020, 13, 3–10.
- Sullivan, M.J.P.; Lewis, S.L.; Affum-Baffoe, K.; Castilho, C.; Costa, F.; Sanchez, A.C.; Ewango, C.E.N.; Hubau, W.; Marimon, B.; Monteagudo-Mendoza, A.; et al. Long-term thermal sensitivity of Earth’s tropical forests. Science 2020, 368, 869–874.
- Cortes, A.J.; Restrepo-Montoya, M.; Bedoya-Canas, L.E. Modern strategies to assess and breed forest tree adaptation to changing climate. Front. Plant Sci. 2020, 11, 583323.
- Neale, D.B.; Kremer, A. Forest tree genomics: Growing resources and applications. Nat. Rev. Genet. 2011, 12, 111–122.
- White, T.L.; Adams, W.T.; Neale, D.B. Forest Genetics; Cabi: Wallingford, UK, 2007.
- Seehausen, O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004, 19, 198–207.
- Burkhart, H.E.; Brunner, A.M.; Stanton, B.J.; Shuren, R.A.; Amateis, R.L.; Creighton, J.L. An assessment of potential of hybrid poplar for planting in the Virginia Piedmont. New For. 2017, 48, 479–490.
- Cipollini, M.; Dingley, N.R.; Felch, P.; Maddox, C. Evaluation of phenotypic traits and blight-resistance in an American chestnut backcross orchard in Georgia. Glob. Ecol. Conserv. 2017, 10, 1–8.
- Badenes, M.L.; Fernández, I.; Martí, A.; Ríos, G.; Rubio-Cabetas, M.J. Application of genomic technologies to the breeding of trees. Front. Genet. 2016, 7, 198.
- Lebedev, V.G.; Lebedeva, T.N.; Chernodubov, A.I.; Shestibratov, K.A. Genomic selection for forest tree improvement: Methods, achievements and perspectives. Forests 2020, 11, 1190.
- Thistlethwaite, F.R.; Ratcliffe, B.; Klápště, J.; Porth, I.; Chen, C.; Stoehr, M.U.; El-Kassaby, Y.A. Genomic selection of juvenile height across a single-generational gap in Douglas-fir. Heredity 2019, 122, 848–863.
- Isik, F.; Bartholomé, J.; Farjat, A.; Chancerel, E.; Raffin, A.; Sanchez, L.; Plomion, C.; Bouffier, L. Genomic selection in maritime pine. Plant Sci. 2016, 242, 108–119.
- Kaiser, N.; Douches, D.; Dhingra, A.; Glenn, K.C.; Herzig, P.R.; Stowe, E.C.; Swarup, S. The role of conventional plant breeding in ensuring safe levels of naturally occurring toxins in food crops. Trends Food Sci. Technol. 2020, 100, 51–66.
- Iwata, H.; Minamikawa, M.F.; Kajiya-Kanegae, H.; Ishimori, M.; Hayashi, T. Genomics-assisted breeding in fruit trees. Breed. Sci. 2016, 66, 100–115.
- Te Beest, M.; Le Roux, J.J.; Richardson, D.M.; Brysting, A.K.; Suda, J.; Kubesova, M.; Pysek, P. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 2012, 109, 19–45.
- Lamo, K.; Bhat, D.J.; Kour, K.; Solanki, S.P.S. Mutation studies in fruit crops: A review. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3620–3633.
- Zonneveld, B.J.M.; Pollock, W.I. Flow cytometric analysis of somaclonal variation in lineages of Hosta sports detects polyploidy and aneuploidy chimeras. Plant Biol. 2012, 14, 972–979.
- Malabarba, J.; Chevreau, E.; Dousset, N.; Veillet, F.; Moizan, J.; Vergne, E. New strategies to overcome present CRISPR/Cas9 limitations in apple and pear: Efficient dechimerization and base editing. Int. J. Mol. Sci. 2020, 22, 319.
- Gambino, G.; Moine, A.; Boccacci, P.; Perrone, I.; Pagliarani, C. Somatic embryogenesis is an effective strategy for dissecting chimerism phenomena in Vitis vinifera cv Nebbiolo. Plant Cell Rep. 2021, 40, 205–211.
- Kumawat, S.; Rana, N.; Bansal, R.; Vishwakarma, G.; Mehetre, S.T.; Das, B.K.; Kumar, M.; Yadav, S.K.; Sonah, H.; Sharma, T.R.; et al. Expanding avenue of fast neutron mediated mutagenesis for crop improvement. Plants 2019, 8, 164.
- Predieri, S. Mutation induction and tissue culture in improving fruits. Plant Cell Tissue Organ Cult. 2001, 64, 185–210.
- Lokko, Y.; Amoatey, H. Improvement of Pineapple Using in Vitro and Mutation Breeding Techniques; Biotechnology and Nuclear Agriculture Research Institute: Legon-Accra, Ghana, 2001; pp. 25–29.
- International Atomic Energy Agency. In Vitro Techniques for Selection of Radiation Induced Mutations Adapted to Adverse Environmental Conditions. In Proceedings of a Final Research Coordination Meeting; Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture: Vienna, Austria, 2001; pp. 1–96.
- Mba, C.; Afza, R.; Bado, S.; Mohan Jain, S. Induced mutagenesis in plants using physical and chemical agents. Plant Cell Cult. Essent. Methods 2010, 20, 111–130.
- Elhiti, M.; Wang, H.Y.; Austin, R.S.; Chen, B.; Brown, D.; Wang, A.M. Generation of chemically induced mutations using in vitro propagated shoot tip tissues for genetic improvement of fruit trees. Plant Cell Tissue Organ Cult. 2016, 124, 447–452.
- Alvarez, D.; Cerda-Bennasser, P.; Stowe, E.; Ramirez-Torres, F.; Capell, T.; Dhingra, A.; Christou, P. Fruit crops in the era of genome editing: Closing the regulatory gap. Plant Cell Rep. 2021, 40, 915–930.
- Sikora, P.; Chawade, A.; Larsson, M.; Olsson, J.; Olsson, O. Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int. J. Plant Genom. 2011, 2011, 314829.
- Kumawat, S.; Rana, N.; Bansal, R.; Vishwakarma, G.; Mehetre, S.; Das, B.K.; Kumar, M.; Yadav, S.; Sonah, H.; Sharma, T.R.; et al. Fast neutron mutagenesis in plants: Advances, applicability and challenges. Plant Sci. 2019.
- Spengler, R.N. Origins of the apple: The role of megafaunal mutualism in the domestication of Malus and rosaceous trees. Front. Plant Sci. 2019, 10, 617.
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, T.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662.
- Crosby, J.A.; Janick, J.; Pecknold, P.C.; Korban, S.S.; Oconnor, P.A.; Ries, S.M.; Goffreda, J.; Voordeckers, A. Breeding apples for scab resistance: 1945–1990. Fruit Var. J. 1992, 46, 145–166.
- Mikulic-Petkovsek, M.; Ivancic, A.; Schmitzer, V.; Veberic, R.; Stampar, F. Comparison of major taste compounds and antioxidative properties of fruits and flowers of different Sambucus species and interspecific hybrids. Food Chem. 2016, 200, 134–140.
- Rajan, R.P.; Singh, G. A review on application of somaclonal variation in important horticulture crops. Plant Cell Biotechnol. Mol. Biol. 2021, 22, 161–175.
- Ranghoo-Sanmukhiya, V.M. Somaclonal variation and methods used for its detection. In Propagation and Genetic Manipulation of Plants; Siddique, I., Ed.; Springer: Singapore, 2021; pp. 1–18.
- Orbović, V.; Ćalović, M.; Viloria, Z.; Nielsen, B.; Gmitter, F.G.; Castle, W.S.; Grosser, J.W. Analysis of genetic variability in various tissue culture-derived lemon plant populations using RAPD and flow cytometry. Euphytica 2008, 161, 329–335.
- Cevallos-Cevallos, J.M.; Jines, C.; Maridueña-Zavala, M.G.; Molina-Miranda, M.J.; Ochoa, D.E.; Flores-Cedeno, J.A. GC-MS metabolite profiling for specific detection of dwarf somaclonal variation in banana plants. Appl. Plant Sci. 2018, 6, e01194.
- Henao-Ramírez, A.M.; Salazar Duque, H.J.; Calle Tobón, A.F.; Urrea Trujillo, A.I. Determination of genetic stability in cacao plants (Theobroma Cacao L.) derived from somatic embryogenesis using microsatellite molecular markers (SSR). Int. J. Fruit Sci. 2021, 21, 284–298.
- Mba, C. Induced mutations unleash the potentials of plant genetic resources for food and agriculture. Agronomy 2013, 3, 200–231.
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeted screening for induced mutations. Nat. Biotechnol. 2000, 18, 455–457.
- Jankowicz-Cieslak, J.; Mba, C.; Till, B.J. Mutagenesis for crop breeding and functional genomics. In Biotechnologies for Plant Mutation Breeding; Springer: Cham, Switzerland, 2017; pp. 3–18.
- Sato, Y.; Sato, K.; Nishio, T. Interspecific pairs of class II S haplotypes having different recognition specificities between Brassica oleracea and Brassica rapa. Plant Cell Physiol. 2006, 47, 340–345.
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstädler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 2017, 15, 367–378.
- Tsai, H.; Howell, T.; Nitcher, R.; Missirian, V.; Watson, B.; Ngo, K.J.; Lieberman, M.; Fass, J.; Uauy, C.; Tran, R.K.; et al. Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol. 2011, 156, 1257–1268.
- Taheri, S.; Abdullah, T.L.; Jain, S.M.; Sahebi, M.; Azizi, P. TILLING, high-resolution melting (HRM), and next-generation sequencing (NGS) techniques in plant mutation breeding. Mol. Breed. 2017, 37, 40.
- Gilchrist, E.J.; Sidebottom, C.H.D.; Koh, C.S.; MacInnes, T.; Sharpe, A.G.; Haughn, G.W. A mutant Brassica napus (Canola) population for the identification of new genetic diversity via TILLING and next generation sequencing. PLoS ONE 2013, 8, e84303.
- Chen, L.; Hao, L.; Parry, M.A.J.; Phillips, A.L.; Hu, Y.-G. Progress in TILLING as a tool for functional genomics and improvement of crops. J. Integrat. Plant Biol. 2014, 56, 425–443.
- Freeman, J.S.; Potts, B.M.; Downes, G.M.; Pilbeam, D.; Thavamanikumar, S.; Vaillancourt, R.E. Stability of quantitative trait loci for growth and wood properties across multiple pedigrees and environments in Eucalyptus globulus. New Phytol. 2013, 198, 1121–1134.
- Osakabe, Y.; Sugano, S.S.; Osakabe, K. Genome engineering of woody plants: Past, present and future. J. Wood Sci. 2016, 62, 217–225.
- Chen, X.; Li, S.; Zhang, D.; Han, M.; Jin, X.; Zhao, C.; Wang, S.; Xing, L.; Ma, J.; Ji, J.; et al. Sequencing of a wild apple (Malus baccata) genome unravels the differences between cultivated and wild apple species regarding disease resistance and cold tolerance. G3-Genes Genom. Genet. 2019, 9, 2051–2060.
- Torales, S.L.; El Mujtar, V.; Marcucci-Poltri, S.; Pomponio, F.; Soliani, C.; Villalba, P.; Estravis-Barcala, M.; Klein, L.; García, M.; Pentreath, V.; et al. Application of high-throughput sequencing technologies in native forest tree species in Argentina: Implications for breeding. In Low Intensity Breeding of Native Forest Trees in Argentina: Genetic Basis for their Domestication and Conservation; Pastorino, M.J., Marchelli, P., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 455–482.
- Ricroch, A.E.; Henard-Damave, M.C. Next biotech plants: New traits, crops, developers and technologies for addressing global challenges. Crit. Rev. Biotechnol. 2016, 36, 675–690.
- Yabor, L.; Perez, L.; Gomez, D.; Villalobos-Olivera, A.; Mendoza, J.R.; Martinez, J.; Escalante, D.; Garro, G.; Hajari, E.; Lorenzo, J.C. Histological evaluation of pineapple transgenic plants following 8 years of field growth. Euphytica 2020, 216, 23.
- Ault, K.; Viswanath, V.; Jayawickrama, J.; Ma, C.; Eaton, J.; Meilan, R.; Beauchamp, G.; Hohenschuh, W.; Murthy, G.; Strauss, S.H. Improved growth and weed control of glyphosate-tolerant poplars. New For. 2016, 47, 653–667.
- Gonsalves, D.; Tripathi, S.; Carr, J.B.; Suzuki, J.Y. Papaya ringspot virus. Plant Health Inst. 2010, 10, 1094.
- Carter, N. Petition for Determination of Nonregulated Status: Arctic™ Apple (Malus × domestica) Events GD743 and GS784; United States Department of Agriculture—Animal and Plant Health Inspection Service: Summerland, BC, Canada, 2012.
- Stowe, E.; Dhingra, A. Development of the Arctic® Apple. Plant Breed. Rev. 2021, 44, 273–296.
- Scorza, R.; Callahan, A.; Ravelonandro, M.; Braverman, M. Development and regulation of the Plum pox virus resistant transgenic plum “HoneySweet”. In Regulation of Agricultural Biotechnology: The United States and Canada; Springer: Berlin/Heidelberg, Germany, 2012; pp. 269–280.
- Xu, K. An overview of Arctic apples: Basic facts and characteristics. NY Fruit Q. 2013, 21, 8–10.
- Food and Drug Administration (FDA). Biotechnology Consultations on Food from GE Plant Varieties; Food and Drug Administration (FDA): Silver Spring, MD, USA, 2017.
- Hu, L.; Lu, H.; Liu, Q.; Chen, X.; Jiang, X. Overexpression of mtlD gene in transgenic Populus tomentosa improves salt tolerance through accumulation of mannitol. Tree Physiol. 2005, 25, 1273–1281.
- Li, Y.L.; Su, X.H.; Zhang, B.Y.; Huang, Q.J.; Zhang, X.H.; Huang, R.F. Expression of jasmonic ethylene responsive factor gene in transgenic poplar tree leads to increased salt tolerance. Tree Physiol. 2009, 29, 273–279.
- Randle, M.; Tennant, P. Transgenic Papaya. In Genetically Modified Crops: Current Status, Prospects and Challenges; Kavi Kishor, P.B., Rajam, M.V., Pullaiah, T., Eds.; Springer: Singapore, 2021; Volume 2, pp. 129–160.
- Vadlamudi, T.; Patil, B.L.; Kaldis, A.; Sai Gopal, D.V.R.; Mishra, R.; Berbati, M.; Voloudakis, A. DsRNA-mediated protection against two isolates of Papaya ringspot virus through topical application of dsRNA in papaya. J. Virol. Meth. 2020, 275, 113750.
- Matsunaga, E.; Nanto, K.; Oishi, M.; Ebinuma, H.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Shibata, D.; Shimada, T. Agrobacterium-mediated transformation of Eucalyptus globulus using explants with shoot apex with introduction of bacterial choline oxidase gene to enhance salt tolerance. Plant Cell Rep. 2012, 31, 225–235.
More
