Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Foad Alzoughool and Version 2 by Peter Tang.
Coronaviruses (CoVs) are a large group of viruses common among many animals, including humans. They can cause respiratory illnesses in humans and gastrointestinal illnesses in animals. Under the electron microscope, virions of CoVs have large peplomers that make it look like a crown, hence the name corona, meaning “crown” or “halo. The emerging coronavirus disease (COVID-19) swept across the world, affecting more than 200 countries and territories.
- COVID-19
- SARS-CoV-2
- Coronavirus
1. Introduction
The coronavirus disease 2019 (COVID-19) was first identified in patients with severe respiratory disease in Wuhan, China. The causative agent was a novel coronavirus scientifically named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1][2][1,2]. Since its discovery, more than 2,850,000 cases have been infected, including nearly 200,000 who have died. The worrisome features of COVID-19 are its apparent ability to spread readily and its propensity to cause severe disease in older adults and patients with existing health conditions [3][4][3,4]. Moreover, coronaviruses are well-known to mutate and recombine [5]. The genomic sequence of SARS-CoV-2 has changed since it was first reported. Some scientists believe that these changes have enhanced the virulence of the virus and there are two circulating strains, deadly strain “L”, and less virulent one “S” [6]. Currently, there are no specific antiviral treatments or vaccines available for COVID-19. Treatments are mainly focusing on symptomatic and respiratory support according to protocols issued by the health authority in each country, where many countries are following WHO protocol [3][4][3,4]. Nearly all cases with severe symptoms accept oxygen. In critical cases and life-threatening situations, passive immunization through the convalescent plasma and immunoglobulin G transfusion can be used as a rescue treatment [7]. Given the current situation, finding a treatment has become a global public health priority. Many companies are working on finding a medication or a viable vaccine for SARS-CoV-2. However, this is not a short process; scientists estimate the period to have one, about 12–18 months.
The COVID-19 is the third novel coronavirus to cause a large-scale epidemic in the twenty-first century after the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) in 2003 [8][9][10][8,9,10] and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012 [11][12][13][11,12,13].
2. Coronaviruses
Coronaviruses (CoVs) are a large group of viruses common among many animals, including humans. They can cause respiratory illnesses in humans and gastrointestinal illnesses in animals. Under the electron microscope, virions of CoVs have large peplomers that make it look like a crown, hence the name corona, meaning “crown” or “halo” [14][15][16][16,17,18]. Before 2003, human CoVs were not considered a deadly virus. The circulating strains were causing mild symptoms in immunocompetent people. Typically, coronavirus symptoms include runny nose, cough, sore throat, headache, and fever that can last for several days. However, in immunocompromised patients, there is a chance that the virus could cause a lower respiratory illness like pneumonia and bronchitis [14][15][16,17]. In 2003, the world was shocked by the first pandemic of the 21st century; the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) emerged in Guangdong, China, resulting in 774 deaths and more than 8000 patients [14][17][18][16,19,20]. Nine years later, a strain of CoV evolved in Saudi Arabia to cause the Middle East Respiratory Syndrome Coronavirus (MERS-CoV), approximately 2500 cases have been confirmed, including 861 deaths with a fearful case–fatality rate of 34.4% [18][20].2.1. Coronaviruses Diversity
CoVs members belong to the subfamily Coronovirinae within the family Coronaviridae and the order Nidovirales. Based on their protein sequences and phylogenetic relationships, members of the Coronavirinae subfamily can be classified into four groups, Alphacoronaviruses, Betacoronaviruses, Gammacoronaviruses, and Deltacoronaviruses. Gammacoronaviruses and Deltacoronaviruses infect birds and might infect mammals, but never reported to cause any illnesses in humans [18][19][20,21]. On the other hand, Alphacoronaviruses and Betacoronaviruses are capable of causing respiratory illnesses in humans and gastrointestinal illnesses in animals. Before December 2019, six common coronaviruses (members of Alphacoronaviruses and Betacoronaviruses) were known to infect humans. HCoV-229E, and HCoV-NL63 and they belong to the Alphacoronaviruses. HCoV-OC43, and HCoV-HKU1 belong to lineage A Betacoronaviruses, and the two deadly viruses SARS-CoV and MERS-CoV belong to lineages B and C of Betacoronaviruses, respectively [18][20]. The genomic analysis found that SARS-CoV-2 belongs to the Betacoronavirus group, lineage B [4][5][4,5]. CoVs are zoonotic pathogens originating in animals and can be transmitted to humans through direct contact. All CoVs that caused epidemics (including COVID-19) are believed to be originated in bats. Bats are hosts of many coronaviruses [15][20][17,22]. However, in most cases, these viruses were transmitted to humans through an intermediate animal host. SARS-CoV started through direct contact with market civets cats [21][23]. MERS-CoV transmitted directly to humans from dromedary camels [11][12][13][11,12,13]. The COVID-19 is suspected to be emerged in the seafood market in Wuhan, China, [1][18][1,20]. Most of the early reported cases have been in that market, which was closed later by the Chinese authority. Evolutionary analysis of COVID-19 virus revealed that it is most similar to the bat SARS-like coronaviruses, and for the similarity, it was named SARS-CoV-2. In summary, most of the scientific report believes that SARS-CoV-2 was originated in bats and transmitted to humans through intermediate animal host in the seafood market [5]. Nevertheless, researchers are yet to find a definitive answer to which animal serves as an intermediate host.2.2. Coronavirus Genome Structure and Replication
The CoVs genome is a single-stranded positive-sense RNA (+ssRNA) molecule. The genome size ranges between 27–32 kbp, one of the largest known RNA viruses (Figure 1) [18][22][20,24]. The genomic structure of CoVs contains at least six open reading frames (ORFs). The first ORFs (ORF1a/b), located at the 5′ end, about two-thirds of the whole genome length, and encodes a polyprotein1a,b (pp1a, pp1b) [23][25]. Other ORFs are located on 3′ end encodes at least four structural proteins: envelop glycoprotein spike (S), responsible for recognizing host cell receptors [24][26]. Membrane (M) proteins, responsible for shaping the virions [25][27]. The envelope (E) proteins, responsible for virions assembly and release [26][28]. The nucleocapsid (N) proteins are involved in packaging the RNA genome and in the virions and play roles in pathogenicity as an interferon (IFN) inhibitor [27][28][29,30]. In addition to the four main structural proteins, there are structural and accessory proteins that are species-specific, such as HE protein, 3a/b protein, and 4a/b protein [22][24]. Once the viral genome enters the cytoplasm of the target cell, and given it is a positive-sense RNA genome, it translates into two polyproteins 1a, b (pp1a, pp1b). These polyproteins are processed into 16 non-structural proteins (NSPs) to form a replication-transcription complex (RTC) that is involved in genome transcription and replication. Consequently, a nested set of subgenomic RNAs (sgRNAs) is synthesized by RTC in the form of discontinuous transcription [22][24].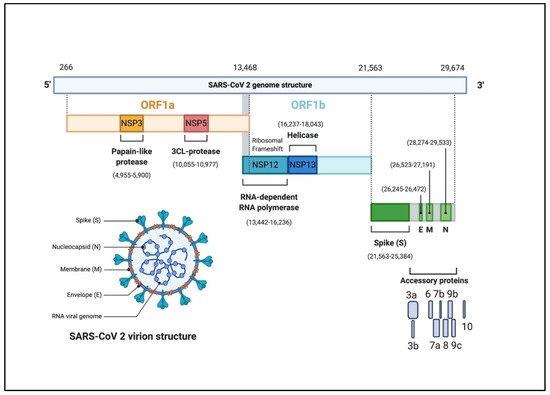
Figure 1. The genomic organization of SARS-CoV-2. The genome encodes two large genes ORF1a (yellow), ORF1b (blue), which encode 16 non-structural proteins (NSP1– NSP16). These NSPs are processed to form a replication–transcription complex (RTC) that is involved in genome transcription and replication. For example, NSP3 and NSP5 encode for Papain-like protease (PLP) and 3CL-protease, respectively. Both proteins function in polypeptides cleaving and block the host innate immune response. NSP12 encodes for RNA-dependent RNA polymerase (RdRp). NSP15 encodes for RNA helicase. The structural genes encode the structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), highlighted in green. The accessory proteins (shades of grey) are unique to SARS-CoV-2 in terms of number, genomic organization, sequence, and function (figure created with biorender.com).
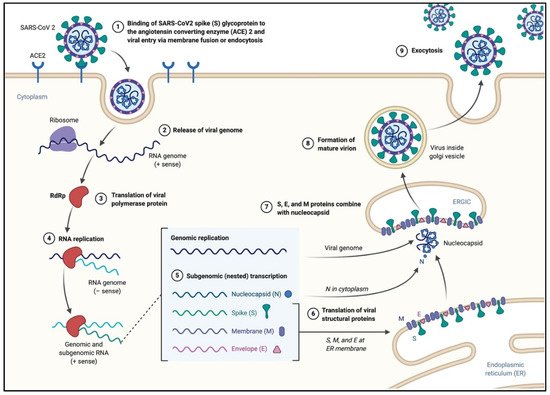
Figure 2. The life cycle of SARS-CoV-2 in the host cells. The S glycoproteins of the virion bind to the cellular receptor angiotensin-converting enzyme 2 (ACE2) and enters target cells through an endosomal pathway. Following the entry of the virus into the host cell, the viral RNA is unveiled in the cytoplasm. ORF1a and ORF1ab are translated to produce pp1a and pp1ab polyproteins, which are cleaved by the proteases of the RTC. During replication, RTC drives the production full length (−) RNA copies of the genome and used as templates for full-length (+) RNA genomes. During transcription, a nested set of sub-genomic RNAs (sgRNAs), is produced in a manner of discontinuous transcription (fragmented transcription). Even though these sgRNAs may have several open reading frames (ORFs), only the closest ORF (to the 5′ end) will be translated. Following the production of SARS-CoV-2 structural proteins, nucleocapsids are assembled in the cytoplasm and followed by budding into the lumen of the endoplasmic reticulum (ER)–Golgi intermediate compartment. Virions are then released from the infected cell through exocytosis (figure created with biorender.com).
