It is well known that correct identification of recovered Aeromonas strains at the genus and species level is a complex process. Biochemical or phenotypic identification tests and specially those included in miniaturized and/or semi-automatic commercial identification systems (API, Vitek, BBLCrystal, MicroScan etc) produce confusion with the Vibrio genus and an erroneous overestimation of the species Aeromonas hydrophila. Correct identification requires the use of molecular techniques, like the detection of the gene that encode for the GCAT (glycerophospholopid-cholesterol acyltransferase) that can discriminate the genus or the analysis of the sequences of housekeeping genes (gyrB, rpoD, etc) to correctly identifying the species. The latter genes are necessary because the 16S rRNA gene does not show enough resolution to discriminate closely related species (i.e. A. salmonicida from A. bestiarum). In fact many new species were discovered thanks to the use of gyrB and rpoD genes for identification, and the construction of a multilocus phylogenetic analysis with the concatenated sequences of five housekeeping genes was used as a tool in their descriptions. The progress in the techniques used to obtain bacteria genomes had an spectacular impact on the genus Aeromonas because the genome of the type strain of the different species are available at the GenBank. Tools developed for bacterial identification based on the comparison of genomes like the in silico DNA-DNA hybridization (isDDH) and the Average Nucleotide Identity (ANI) provides objective criteria to define if two genomes belong or not to the same species. This review aims to guide microbiologists in the correct identification of the Aeromonas spp.
- Aeromonas
- genomics
- housekeeping genes
- Aeromonas, housekeeping genes, genomics.
1. Introduction
For the description of new prokaryotic species, the International Committee on Systematics of Prokaryotes (ICSP) recommends a polyphasic study, which should include phenotypic and phylogenetic differentiation from existing species[1]. A discussion of the criteria proposed by the ICSP in relation to the genus Aeromonas is given elsewhere[2].
2. Phenotypic Identification
Phenotypic identification is made by physiological, morphological, and biochemical characteristics. Classic phenotypic characteristics [1][3][4] that identify the genus Aeromonas are Gram-negative staining, the presence of normally positive cytochrome oxidase, and growth in nutritive broth at 0% to grow in the presence of vibriostatic factor O/129 [1][3]. Despite that, identification to the species level using this approach is difficult due to the variable behavior of the strains. In 2010, Beaz-Hidalgo et al. [5] re-identified 119 strains, isolated mainly from diseased fish that had previously been identified phenotypically. The re-identification was carried out by molecular methods (16S rRNA-RFLP and rpoD sequences) and the results demonstrated that only 35.5% were correctly identified at the species level.
Additionally, commercial identification systems (API 20E, Vitek, BBL Crystal, MicroScan W/A, among others) have commonly been used in clinical laboratories, although several authors demonstrated that these systems had limitations [6][7]. In 2010, Lamy et al. [7] compared the accuracy of six commercial systems for Aeromonas identification, using the rpoB sequencing as a reference. Concordance was shown to be low between phylogenetic identification and the commercial identifications systems, with erroneous identification at species level. The study also ratified results of previous studies that highlighted the confusion between Aeromonas and the genus Vibrio [7][8][9]. To avoid this confusion a DNA probe, base on the detection of the gene that encode the glycerophospholopid-cholesterol acyltransferase (GCAT) specific for Aeromonas colonies, was developed by Chacón et al. [8]. A misidentification of a clinical strain identified at the hospital as Aeromonas sp. using API 20E and API 20NE, showed to correspond to Vibrio alginolyticus after a molecular identification.[10]
3. Molecular Identification
3.1. Techniques Based on the 16S rRNA Gene
The 16S rRNA gene is considered a stable molecular marker for identifying bacterial species, since its distribution is universal and allows comparison of microorganisms [1][2][11][12]. In addition, its structure presents a mosaic of variable regions, suitable in the differentiation of closely related organisms, and their conserved regions are useful for the distant organisms comparison and this allows for the design of “universal” primers [13].
In 1992, Martínez-Murcia et al. [14] sequenced the 16S rRNA gene for the first time using strains of the species described up to then; the results agreed with the DNA–DNA hybridization (DDH). In the genus Aeromonas, the 16S rRNA gene has an interspecies similarity range from 96.7–100% and the informative nucleotide positions are located mainly on region V3 [12][13].
Additionally, the presence of microheterogeneities (i.e., mutations on specific positions of the sequence of one of several copies of the 16S rRNA gene) in combination with the high similarity of the sequences for closely related species makes this gene not suitable for the Aeromonas spp. identification [13][15][16]. Figure 1 shows the phylogenetic tree derived from sequences of the 16S rRNA gene of the 36 Aeromonas species.
Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) is a powerful method introduced to many clinical laboratories in recent years for the identification and comparison of microbial isolates [10][17]. The MALDI-TOF MS mainly detects proteins associated with the 16S rRNA gene and therefore the low resolution of this gene for the identification of closely related species of Aeromonas also impacts the resolution of this method [18].
In 2014, Chen et al. [19] used the MALDI-TOF MS to characterize 217 clinical isolates previously identified by rpoB sequencing and found that 100% were correctly identified at genus level, and 97% at species level. One-year later, Shin et al. [20] re-identified 65 clinical strains previously identified by gyrB sequencing and showed 98.5% concordance at genus level, and 92.3% at the species level using the MALDI-TOF MS. These results are relatively similar to those reported by Latif-Eugenín [18] who identified 179 clinical strains from Spanish hospitals, with 98.3% correct identification at genus level, and 91.1% at species level using MALDI-TOF MS. Based on those data, they suggested that MALDI-TOF MS is a useful tool, since the identification error was <10%, while with phenotypic identification methods error can be very high. The main limitation of the latter method is the need to update the database to include the many missing Aeromonas species, such as A. dhakensis or the new species (A. intestinalis, A. crassostreae, A. enterica, and A. aquatilis). A recent study that used MALDI-TOF MS for the characterization of Aeromonas strains isolated from fish [21] demonstrated that the number of correct identifications increased after the addition of 14 new spectra in the MALDI-TOF Biotyper database.
3.2. Housekeeping Genes
Housekeeping genes (HKG) encode proteins with essential functions for the survival of bacteria. They were introduced for the description of new species using an MLPA because the resolution is higher than the 16S rRNA gene [1][22]. For taxonomic analysis, the ideal HKG should have the following characteristics: (1) they should not be influenced by horizontal gene transfer; (2) they should be present in all bacteria; (3) they should be single genes in the genome of the bacteria; (4) and finally they should present at least two conserved regions for the design of primers [23].
The first HKG studied of Aeromonas was the gyrB gene that encodes the subunit B of DNA gyrase [24]. Another HKG that shows a similar phylogeny to gyrB is the rpoD gene that encodes sigma factor S70 (that confers promoter-specific transcription initiation for the RNA polymerase)[6]. These genes were used to recognize and describe many species in recent years [25][26][27][28][29][30][31][32][33][34][35][36][37]. The phylogenetic tree derived from sequences of gyrB and rpoD genes of 22 highly similar Aeromonas species based on the 16S rRNA gene is presented in Figure 1. Many studies have described other HKG: rpoB, recA, dnaJ, cpn60, mdh, gyrA, dnaX, atpD, groL, gltA, metG, ppsA, dnaK, radA, tsF, and zipA [16][31][38][39][40][41][42][43]. However, the phylogeny based on the sequence of only one HKG is sometimes not conclusive and a higher resolution is obtained using the concatenated sequences of several HKG [1][2][13]. In 2011, Martínez-Murcia et al.[38] described the first MLPA of the genus Aeromonas using the concatenated sequences of seven genes (rpoD, gyrB, gyrA, atpD, recD, dnaJ, and dnaX).
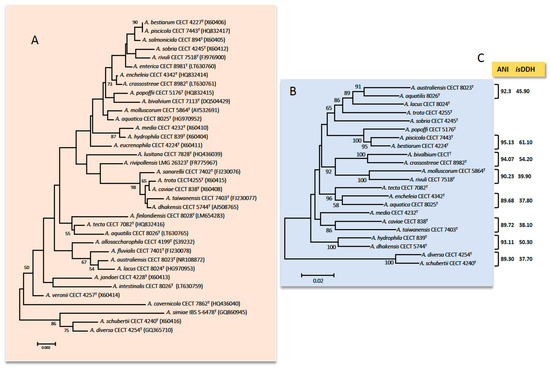
Figure 1. (A) Phylogenetic tree based on the sequences of the 16S rRNA gene (1498 bp) among 36 species of Aeromonas. (B) Phylogenetic tree based on the concatenated sequences of rpoD and gyrB genes (1098) among the most similar species based on the 16S rRNA gene. The number in the nodes indicates the bootstrap values substitutions estimated by site. (C) Results (%) for the ANI (average nucleotide identity) and isDDH (in silico DNA–DNA hybridization) obtained between the genomes of the most similar species; notice that ANI and isDDH values are ≤96% and ≤70% in all cases, respectively which are the cut-off values established for delimiting Aeromonas spp.
3.3. Genotyping Methods
Different molecular methods have been employed to trace whether two isolates of Aeromonas belong, or not, to the same clone and therefore share an epidemiological relationship [10][38]. These methods are the enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR), the randomly amplified polymorphic DNA-PCR (RAPD-PCR), the amplified fragment length polymorphism (AFLP), the pulsed-field gel electrophoresis (PFGE), and the multilocus sequence typing (MLST) [10][38].
The ERIC-PCR is one of the most popular methods for genotyping Aeromonas because it is easy to carry out, does not require any expensive equipment, and is highly reproducible [44][45]. Consequently, it has been used in several epidemiological studies [46][47][48][49]. In a recent study, one strain of A. caviae isolated from a sample of lettuce showed the same ERIC genotype pattern as a strain recovered from a sample of irrigation water [49]. In addition, the same genotype of A. sanarellii was recovered in samples of parsley and tomato that were irrigated with the same reclaimed water, confirming the potential health risk to humans [49].
The MLST is based on the analysis of the sequences of several genes, normally seven, to recognize allele sequences [50]. This technique show to be highly discriminatory and reproducible compared with other techniques, and there is also a database to help investigators compare their results. The Bacterial Isolate Genome Sequence Database (BIGSdb) is the platform that currently manages the MLST database [50] and can be found within the PubMLST public databases. The MLST scheme is freely available and was created for Aeromonas in 2010 based on the data obtained by Martino et al. [42] using six genes (gyrB, groL, gltA, metG, ppsA, and recA). The major problem of the latter technique is the need for perfect sequences of seven housekeeping genes (450–500 bp each gene) with no ambiguities, and result comparison might be limited by the number of strains and origins available in the database, which in its last update (22 October 2019) had 751 strains, and 2817 sequences that corresponded to 652 MLST profiles (available online: https://pubmlst.org/Aeromonas/submission.shtml consulted on 22 October 2019).
3.4. Genomics
As of 2 September 2019, 410 Aeromonas genomes were made publicly available in the GenBank database, of which 63 are complete (available online: https://www.ncbi.nlm.nih.gov/genome/?term=Aeromonas). The size of Aeromonas genomes varies between 3.90 Mbp (A. fluvialis) and 5.18 Mbp (A. piscicola) with an average size of 4.51 Mbp [51]. Furthermore, the percentage of G + C was 60.2%, varying between 58.1% (A. australiensis) and 62.8% (A. taiwanensis).
Advances in methods of obtaining complete genomes have increased the number of available genomes in recent years. In fact, only six Aeromonas genomes were available in 2012 [52][53][54][55][56][57] and just two years later, that increased to 56 genomes representing 29 recognized or proposed species of the genus Aeromonas [58]. In 2015, using MLPA and pairwise comparison using the average nucleotide identity (ANI), Beaz-Hidalgo et al. [59] re-identified 44 genomes that were deposited in GenBank, demonstrating that 14 were wrongly labeled by using the MLPA and pairwise comparison using the ANI. The data obtained in that study showed the importance of verifying the taxonomic position of a genome, using the mentioned tools (MLPA and ANI) before submission to the NCBI or other databases. These misidentifications might also be determined using another tool based on genome comparison (i.e., in silico DNA–DNA hybridization (isDDH)) [4][58][59].
The experimental DDH is commonly used for species delineation [35]. However, this technique produces errors and takes up a lot of time. The isDDH described by Kolthoff et al. [60], using the genome-to-genome distance calculator (GGDC) developed by DSMZ (Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany) showed to be an excellent tool for determining the genetic similarity between two bacteria genomes. The results ≥70% indicates that these two strains belong to the same species (Figure 1). Moreover, in 2009, Richter and Rosselló-Mora [61] defined the ANI as the percentage of identity that can be found in the nucleotide sequences of orthologous genes common in the two genomes (Figure 1). Based on different studies [61][62] the cut-off value was established at 95–96% and the results agreed with isDDH. There are several tools to calculate ANI values: JSpecies, ANI calculator, OrthoANI, and OrthoANI-usearch tool. The cut-off for Aeromonas was established in 2014 by Colston et al. [58]. In the study, 56 Aeromonas genomes were analyzed, suggesting that values ≥96% indicate that the two strains belong to different species. Figueras et al. [63] indicated that the ANI and the MLPA are excellent tools for verifying the identity of genomes before they are deposited in GenBank, which would prevent them from being mislabeled. In fact, Beaz-Hidalgo et al. [59] used these tools for re-identifying the genomes deposited in the NCBI, and found that 35.9% of the genomes of non-type strains of Aeromonas spp. were incorrectly labeled. Recently, different studies used the genomic indices to increase the correct identification of ambiguous Aeromonas strains, supporting the notion that these methods are essential in taxonomy [64][65][66].
References
- Stackebrandt, E.; Garrity, G.M.; Trüper, H.G.; Whitman, W.B.; Grimont, P.A.D.; Nesme, X.; Frederiksen, W.; Vauterin, L.; Kampfer, P.; Rosselló-Mora, R.; et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 2002, 52, 1043–1047.
- Figueras, M.J.; Beaz-Hidalgo, R.; Collado, L.; Martínez-Murcia, A.J. Recommendations for a new bacterial species description based on analysis of the unrelated genera Aeromonas and Arcobacter. Bull. BISMiS 2011, 2, 1–16.
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73.
- Abbott, S.L.; Cheung, W.K.W.; Janda, J.M. The genus Aeromonas: Biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 2003, 41, 2348–2357.
- Beaz-Hidalgo, R.; Alperi, A.; Buján, N.; Romalde, J.L.; Figueras, M.J. Comparison of phenotypical and genetic identification of Aeromonas strains isolated from diseased fish. Syst. Appl. Microbiol. 2010, 33, 149–153.
- Soler, L.; Yáñez, M.A.; Chacon, M.R.; Aguilera-Arreola, M.G.; Catalan, V.; Figueras, M.J.; Martínez-Murcia, A.J. Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int. J. Syst. Evol. Microbiol. 2004, 54, 1511–1519.
- Lamy, B.; Laurent, F.; Verdier, I.; Decousser, J.-W.; Lecaillon, E.; Marchandin, H.; Roger, F.; Tigaud, S.; De Montclos, H.; Kodjo, A. Accuracy of 6 commercial systems for identifying clinical Aeromonas isolates. Diagn. Microbiol. Infect. Dis. 2010, 67, 9–14.
- Chacón, M.R.; Castro-Escarpulli, G.; Soler, L.; Guarro, J.; Figueras, M.J. A DNA probe specific for Aeromonas colonies. Diagn. Microbiol. Infect. Dis. 2002, 44, 221–225.
- Soler, L.; Marco, F.; Vilá, J.; Chacon, M.R.; Guarro, J.; Figueras, M.J. Evaluation of two miniaturized systems, MicroScan W/A and BBL Crystal E/NF, for identification of clinical isolates of Aeromonas spp. J. Clin. Microbiol. 2003, 41, 5732–5734.
- Fernández-Bravo, A. Epidemiology and pathogenic characterization of species of the genus Aeromonas. PhD Thesis, Universidad Rovira i Virgili: Tarragona, Spain, 2019.
- Woo, P.; Lau, S.; Teng, J.; Tse, H.; Yuen, K.-Y. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 2008, 14, 908–934.
- Vega-Sánchez, V. Caracterización fenotípica y genotípica de aislamientos de Aeromonas spp. obtenidos de trucha arcoíris (Oncorhynchus mykiss); Universidad Autónoma del Estado de México: Toluca, Mexico, 2015.
- Martínez-Murcia, A.; Lamy, B. Molecular diagnostics by genetic methods. In Aeromonas; Academic Press: Norfolk, UK, 2015; pp. 155–200.
- Martínez-Murcia, A.J.; Benlloch, S.; Collins, M.D. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: Lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 1992, 42, 412–421.
- Alperi, A.; Figueras, M.J.; Inza, I.; Martínez-Murcia, A.J. Analysis of 16S rRNA gene mutations in a subset of Aeromonas strains and their impact in species delineation. Int. Microbiol. 2008, 11, 185–194.
- Roger, F.; Marchandin, H.; Jumas-Bilak, E.; Kodjo, A.; Lamy, B. Multilocus genetics to reconstruct aeromonad evolution. BMC Microbiol. 2012, 12, 62.
- Vavrova, A.; Balazova, T.; Sedlacek, I.; Tvrzova, L.; Sedo, O. Evaluation of the MALDI-TOF MS profiling for identification of newly described Aeromonas spp. Folia Microbiol. 2015, 60, 375–383.
- Latif-Eugenin, F.L. Aeromonas, un microorganismo ambiental de importancia en salud humana y animal. PhD Thesis Universidad Rovira i Virgili: Tarragona, Spain, 2015.
- Chen, P.-L.; Wu, C.-J.; Chen, C.-S.; Tsai, P.-J.; Tang, H.-J.; Ko, W.-C.; Cutler, S. A comparative study of clinical Aeromonas dhakensis and Aeromonas hydrophila isolates in southern Taiwan: A. dhakensis is more predominant and virulent. Clin. Microbiol. Infect. 2014, 20, O428–O434.
- Shin, H.B.; Yoon, J.; Lee, Y.; Kim, M.S.; Lee, K. Comparison of MALDI-TOF MS, housekeeping gene sequencing, and 16S rRNA gene sequencing for identification of Aeromonas clinical isolates. Yonsei Med. J. 2015, 56, 550–555.
- Pérez-Sancho, M.; Cerdá, I.; Fernández-Bravo, A.; Domínguez, L.; Figueras, M.J.; Fernández-Garayzábal, J.F.; Vela, A.I. Limited performance of MALDI-TOF for identification of fish Aeromonas isolates at species level. J. Fish Dis. 2018, 41, 1485–1493.
- Zhong, C.; Han, M.; Yang, P.; Chen, C.; Yu, H.; Wang, L.; Ning, K. Comprehensive analysis reveals the evolution and pathogenicity of Aeromonas, viewed from both single isolated species and microbial communities. mSystems 2019, 4.
- Harayama, S.; Kasai, H. Bacterial phylogeny reconstruction from molecular sequences. In Molecular Identification, Systematics, and Population Structure of Prokaryotes; Springer Science and Business Media: Berlin/Heidelberg, Germany, 2006; pp. 105–139.
- Yáñez, M.A.; Catalan, V.; Apráiz, D.; Figueras, M.J.; Martínez-Murcia, A.J. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 2003, 53, 875–883.
- Martínez-Murcia, A.; Beaz-Hidalgo, R.; Navarro, A.; Carvalho, M.J.; Aravena-Román, M.; Correia, A.; Figueras, M.J.; Saavedra, M.J. Aeromonas lusitana sp. nov., isolated from untreated water and vegetables. Curr. Microbiol. 2016, 72, 795–803.
- Beaz-Hidalgo, R.; Latif-Eugenin, F.; Hossain, M.; Berg, K.; Niemi, R.; Rapala, J.; Lyra, C.; Liles, M.; Figueras, M.J. Aeromonas aquatica sp. nov., Aeromonas finlandiensis sp. nov. and Aeromonas lacus sp. nov. isolated from Finnish waters associated with cyanobacterial blooms. Syst. Appl. Microbiol. 2015, 38, 161–168.
- Figueras, M.J.; Latif-Eugenin, F.; Ballester, F.; Pujol, I.; Tena, D.; Berg, K.; Hossain, M.J.; Beaz-Hidalgo, R.; Liles, M.R. ‘Aeromonas intestinalis’ and ‘Aeromonas enterica’ isolated from human faeces, ‘Aeromonas crassostreae’ from oyster and ‘Aeromonas aquatilis’ isolated from lake water represent novel species. New Microbes New Infect. 2017, 15, 74–76.
- Aravena-Román, M.; Beaz-Hidalgo, R.; Inglis, T.J.J.; Riley, T.V.; Martínez-Murcia, A.J.; Chang, B.J.; Figueras, M.J. Aeromonas australiensis sp. nov., isolated from irrigation water. Int. J. Syst. Evol. Microbiol. 2013, 63, 3130.
- Martinez-Murcia, A.; Beaz-Hidalgo, R.; Svec, P.; Saavedra, M.J.; Figueras, M.J.; Sedlacek, I. Aeromonas cavernicola sp. nov., isolated from fresh water of a brook in a cavern. Curr. Microbiol. 2013, 66, 197–204.
- Beaz-Hidalgo, R.; Martinez-Murcia, A.; Figueras, M.J. Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al. 2002 and Aeromonas aquariorum Martinez-Murcia et al. 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila. Syst. Appl. Microbiol. 2013, 36, 171–176.
- Miñana-Galbis, D.; Farfán, M.; Lorén, J.G.; Fusté, M.C. Proposal to assign Aeromonas diversa sp. nov. as a novel species designation for Aeromonas group 501. Syst. Appl. Microbiol. 2010, 33, 15–19.
- Alperi, A.; Martínez-Murcia, A.J.; Monera, A.; Saavedra, M.J.; Figueras, M.J. Aeromonas fluvialis sp. nov., isolated from a Spanish river. Int. J. Syst. Evol. Microbiol. 2010, 60, 1008.
- Beaz-Hidalgo, R.; Alperi, A.; Figueras, M.J.; Romalde, J.L. Aeromonas piscicola sp. nov., isolated from diseased fish. Syst. Appl. Microbiol. 2009, 32, 471–479.
- Marti, E.; Balcázar, J.L. Aeromonas rivipollensis sp. nov., a novel species isolated from aquatic samples. J. Basic Microbiol. 2015, 55, 1435–1439.
- Figueras, M.J.; Alperi, A.; Beaz-Hidalgo, R.; Stackebrandt, E.; Brambilla, E.; Monera, A.; Martinez-Murcia, A.J. Aeromonas rivuli sp. nov., isolated from the upstream region of a karst water rivulet. Int. J. Syst. Evol. Microbiol. 2011, 61, 242–248.
- Alperi, A.; Martinez-Murcia, A.J.; Ko, W.C.; Monera, A.; Saavedra, M.J.; Figueras, M.J. Aeromonas taiwanensis sp. nov. and Aeromonas sanarellii sp. nov., clinical species from Taiwan. Int. J. Syst. Evol. Microbiol. 2010, 60, 2048–2055.
- Demarta, A.; Küpfer, M.; Riegel, P.; Harf-Monteil, C.; Tonolla, M.; Peduzzi, R.; Monera, A.; Saavedra, M.J.; Martínez-Murcia, A. Aeromonas tecta sp. nov., isolated from clinical and environmental sources. Syst. Appl. Microbiol. 2008, 31, 278–286.
- Martínez-Murcia, A.J.; Monera, A.; Saavedra, M.J.; Oncina, R.; Lopez-Alvarez, M.; Lara, E.; Figueras, M.J. Multilocus phylogenetic analysis of the genus Aeromonas. Syst. Appl. Microbiol. 2011, 34, 189–199.
- Küpfer, M.; Kuhnert, P.; Korczak, B.M.; Peduzzi, R.; Demarta, A. Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. Int. J. Syst. Evol. Microbiol. 2006, 56, 2743–2751.
- Sepe, A.; Barbieri, P.; Peduzzi, R.; Demarta, A. Evaluation of recA sequencing for the classification of Aeromonas strains at the genotype level. Lett. Appl. Microbiol. 2008, 46, 439–444.
- Miñana-Galbis, D.; Urbizu-Serrano, A.; Farfán, M.; Fusté, M.C.; Lorén, J.G. Phylogenetic analysis and identification of Aeromonas species based on sequencing of the cpn60 universal target. Int. J. Syst. Evol. Microbiol. 2009, 59, 1976–1983.
- Martino, M.E.; Fasolato, L.; Montemurro, F.; Rosteghin, M.; Manfrin, A.; Patarnello, T.; Novelli, E.; Cardazzo, B. Determination of microbial diversity of Aeromonas strains on the basis of multilocus sequence typing, phenotype, and presence of putative virulence genes. Appl. Environ. Microbiol. 2011, 77, 4986–5000.
- Nhung, P.H.; Hata, H.; Ohkusu, K.; Noda, M.; Shah, M.M.; Goto, K.; Ezaki, T. Use of the novel phylogenetic marker dnaJ and DNA-DNA hybridization to clarify interrelationships within the genus Aeromonas. Int. J. Syst. Evol. Microbiol. 2007, 57, 1232–1237.
- Figueras, M.J.; Suarez-Franquet, A.; Chacon, M.R.; Soler, L.; Navarro, M.; Alejandre, C.; Grasa, B.; Martínez-Murcia, A.J.; Guarro, J. First record of the rare species Aeromonas culicicola from a drinking water supply. Appl. Environ. Microbiol. 2005, 71, 538–541.
- Soler, L.; Figueras, M.J.; Chacón, M.R.; Guarro, J.; Martinez-Murcia, A.J. Comparison of three molecular methods for typing Aeromonas popoffii isolates. Antonie Leeuwenhoek 2003, 83, 341–349.
- Aguilera-Arreola, M.G.; Hernandez-Rodriguez, C.; Zuniga, G.; Figueras, M.J.; Castro-Escarpulli, G.; Hernández-Rodríguez, C. Aeromonas hydrophila clinical and environmental ecotypes as revealed by genetic diversity and virulence genes. FEMS Microbiol. Lett. 2005, 242, 231–240.
- Maiti, N.; Mandal, A.; Mohanty, S.; Mandal, R. Phenotypic and genetic characterization of Edwardsiella tarda isolated from pond sediments. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 1–8.
- Fontes, M.C.; Saavedra, M.J.; Monera, A.; Martins, C.; Martínez-Murcia, A. Phylogenetic identification of Aeromonas simiae from a pig, first isolate since species description. Veter. Microbiol. 2010, 142, 313–316.
- Latif-Eugenín, F.; Beaz-Hidalgo, R.; Figueras, M.J. Evaluation of different conditions and culture media for the recovery of Aeromonas spp. from water and shellfish samples. J. Appl. Microbiol. 2016, 121, 883–891.
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595.
- Beaz-Hidalgo, R.; Figueras, M.J. Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish Dis. 2013, 36, 371–388.
- Seshadri, R.; Joseph, S.W.; Chopra, A.K.; Sha, J.; Shaw, J.; Graf, J.; Haft, D.; Wu, M.; Ren, Q.; Rosovitz, M.J.; et al. Genome sequence of Aeromonas hydrophila ATCC 7966T: Jack of all trades. J. Bacteriol. 2006, 188, 8272–8282.
- Charette, S.J.; Brochu, F.; Boyle, B.; Filion, G.; Tanaka, K.H.; Derome, N. Draft genome sequence of the virulent strain 01-B526 of the fish pathogen Aeromonas salmonicida. J. Bacteriol. 2012, 194, 722–723.
- Li, Y.; Liu, Y.; Zhou, Z.; Huang, H.; Ren, Y.; Zhang, Y.; Li, G.; Zhou, Z.; Wang, L. Complete genome sequence of Aeromonas veronii strain B565. J. Bacteriol. 2011, 193, 3389–3390.
- Reith, M.E.; Singh, R.K.; Curtis, B.; Boyd, J.M.; Bouevitch, A.; Kimball, J.; Munholland, J.; Murphy, C.; Sarty, D.; Williams, J.; et al. The genome of Aeromonas salmonicida subsp. salmonicida A449: Insights into the evolution of a fish pathogen. BMC Genom. 2008, 9, 427.
- Wu, C.-J.; Wang, H.-C.; Chen, C.-S.; Shu, H.-Y.; Kao, A.-W.; Chen, P.-L.; Ko, W.-C. Genome sequence of a novel human pathogen, Aeromonas aquariorum. J. Bacteriol. 2012, 194, 4114–4115.
- Beatson, S.A.; das Gracas de Luna, M.; Bachmann, N.L.; Alikhan, N.F.; Hanks, K.R.; Sullivan, M.J.; Wee, B.A.; Freitas-Almeida, A.C.; Dos Santos, P.A.; de Melo, J.T.; et al. Genome sequence of the emerging pathogen Aeromonas caviae. J. Bacteriol. 2011, 193, 1286–1287.
- Colston, S.M.; Fullmer, M.S.; Beka, L.; Lamy, B.; Gogarten, J.P.; Graf, J. Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. MBio 2014, 5.
- Beaz-Hidalgo, R.; Hossain, M.J.; Liles, M.R.; Figueras, M.-J. Strategies to avoid wrongly labelled genomes using as example the detected wrong taxonomic affiliation for Aeromonas genomes in the genbank database. PLoS ONE 2015, 10.
- Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Klenk, H.-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418.
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131.
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351.
- Figueras, M.J.; Beaz-Hidalgo, R.; Hossain, M.J.; Liles, M.R. Taxonomic affiliation of new genomes should be verified using average nucleotide identity and multilocus phylogenetic analysis. Genome Announc. 2014, 2.
