Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Saadi Khochbin.
NUT (Nuclear protein in Testis) is a testis-specific factor originally discovered as a chromosomal fusion partner of BRD4 and BRD3, both members of the BET double bromodomain-containing family of proteins, in an aggressive cancer known as NUT Carcinoma (NC).
- NUT midline carcinoma
- NUT carcinoma
1. Introduction
The systematic involvement of NUTM1 in NUT Carcinoma suggested that its encoded protein, NUT could be a central element in the underlying oncogenic molecular mechanisms. However, besides the specific expression of its encoding gene in testis, nothing was known on the physiological and pathological functions of NUT. The presence of members of the BET (Bromodomain and Extra-Termina) family, especially BRD4 (bromodomain containing 4), in fusion with NUT in a large number of NC cancer cases, suggested that the oncogenic activity driven by the BRD4/3-NUT fusion proteins could involve a yet unknown cooperation between the two fusion partners. More specifically, since the proteins of the BET family are known histone/protein acetylation readers, this strong bias towards bromodomain factors among all the identified NUT fusion partners, suggested that histone acetylation and acetylation reading factors could be important components of the underlying oncogenic mechanism [3][1]. Furthermore, because of its physiological context of expression in male germ cells, the hypothesis was made that NUT could also play a role in histone hyperacetylation associated with genome-wide histone-to-protamine replacement, which occurs in the late phases of spermatogenesis [4][2].
Histone-to-protamine replacement is one of the most spectacular known large-scale genome packaging reorganizations known in eukaryotes [5][3]. Despite its dramatic nature and its essential role in procreation and species perpetuation, the molecular basis of this remarkable and unique genome reorganization has remained a black box in biology for many years.
Early investigations, which aimed at deciphering the molecular basis of Histone-to-protamine replacement using mouse models, revealed that the process of histone replacement is associated with the occurrence of a wave of histone hyperacetylation in mice [6][4], as well as in various other species (please see [7][5], and references therein).
After the ground-breaking discovery that bromodomains are binders for acetylated histones by Ming-Ming Zhou and colleagues [8][6], ouresearchers laboratory hypothesized that bromodomain-containing factors could also mediate events which are dependent on histone hyperacetylation in post-meiotic haploid spermatogenic cells (or spermatids) in relation to histone-to-Protamine replacement.
Following this hypothesis, an in silico approach was used to identify putative testis-specific bromodomain-containing factors as possible candidates capable of acting on hyperacetylated histones in spermatids. This strategy identified Brdt (bromodomain testis-specific), a gene encoding a testis-specific member of the BET family, whose function was completely unknown at that time, as a possible candidate [9][7]. Functional investigations that followed revealed the capacity of ectopically expressed BRDT to dramatically compact and reorganize chromatin in response to chromatin hyperacetylation induced by histone deacetylase (HDAC) inhibitor, TSA, treatment [9,10][7][8]. Further structural studies showed that BRDT’s first bromodomain (BD1) presents the remarkable property to specifically bind histone H4 tail bearing simultaneous acetylation at K5 and K8 [11][9]. Since the co-acetylation of these two lysines is a signature of hyperacetylated H4 [12[10][11],13], these studies reenforced ouresearchers hypothesis that BRDT could be a factor acting on hyperacetylated chromatin at the time of histone-to-protamine replacement. This ability of BRDT to bind hyperacetylated H4 was also confirmed in vivo [14][12] and later BD1 was also shown to bind nucleosomal DNA, in addition of H4K5acK8ac, increasing the stability of its interaction with hyperacetylated H4-bearing nucleosomes [15][13]. Finally, by investigating various mouse Brdt genetic models, ouresearchers laboratory confirmed the role of BRDT’s BD1 in the removal and replacement of histones in spermatids [10][8].
However, despite this progress in ouresearchers understanding of the connection between H4 hyperacetylation and histone-to-protamine replacement, the origin of the histone H4 hyperacetylation wave has remained obscure.
In order to obtain a full understanding of this acetylation-dependent histone eviction, it appeared of crucial importance to discover the origin of spermatid-specific H4 hyperacetylation.
Following a series of unsuccessful attempts in ouresearchers laboratory to find mechanisms underlying H4 hyperacetylation in spermatids, weresearchers thought that functional studies of the fusion protein BRD4-NUT in cancer cells could shed some light on the connection between histone acetylation and acetylation-dependent events in spermatids. Indeed, since NUT, a testis-specific factor of unknown function, is fused to and cooperates with BRD4 in the context of NC, the hypothesis was made that, in its physiological context, NUT cooperates with BRDT, and that the NC chromosomal translocation observed in the context of somatic cells cancer actually re-establishes this cooperation.
Hence, functional studies of BRD4-NUT were undertaken [3][1] with the hope that the understanding its function in the context of NC would also provide clues to fully understand the role of histone hyperacetylation in spermatids.
This reasoning turned out to be correct, since these early investigations of the BRD4-NUT fusion protein allowed ouresearchers laboratory to unravel the mechanism by which this functional cooperation between NUT and BRD4 could support the oncogenic activity of the fusion protein. Indeed, the data obtained showed that BRD4-NUT drives chromatin hyperacetylation through a feedforward loop, resulting in the generation of hyperacetylated chromatin foci [3][1]. (Figure 1).
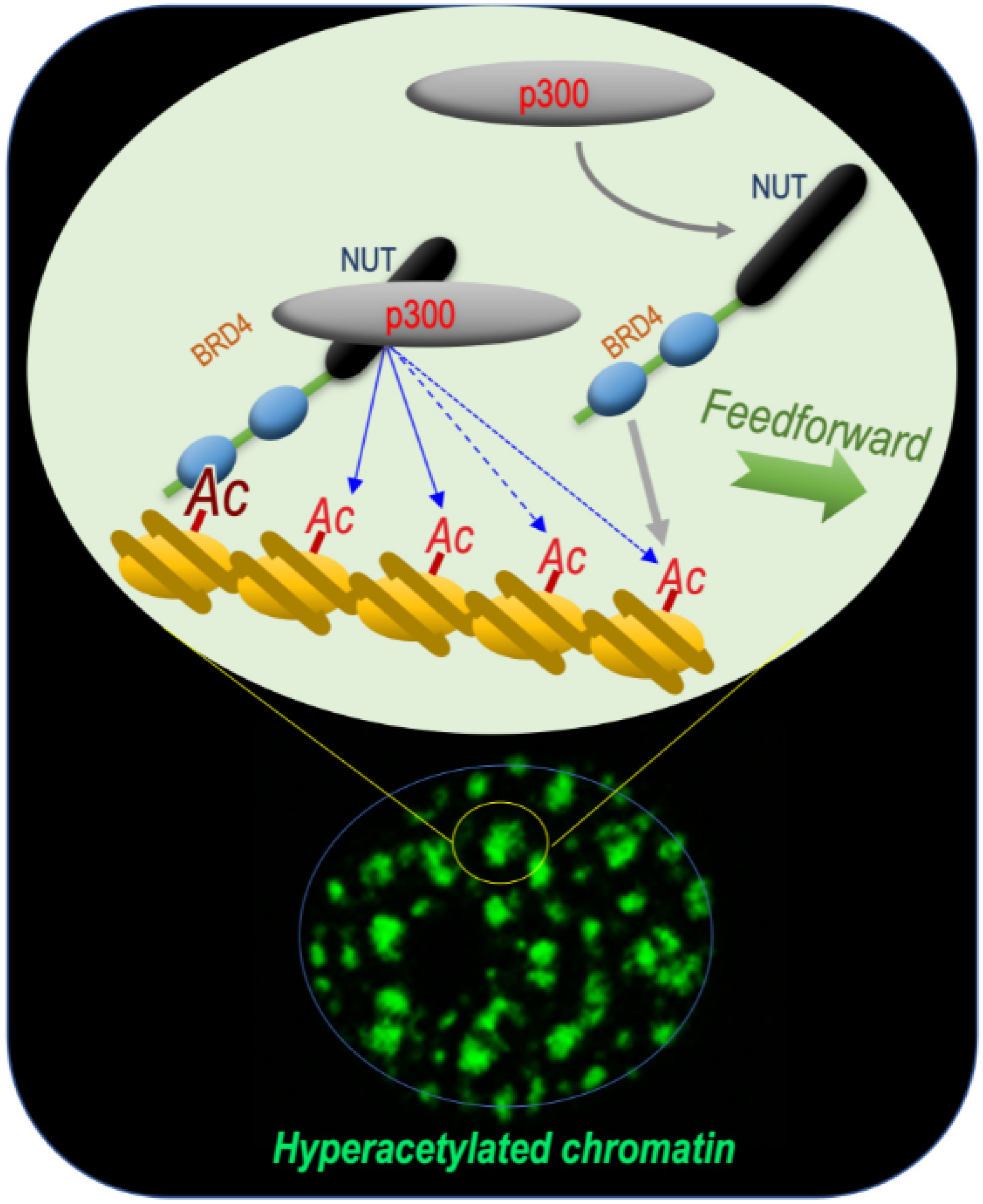
Figure 1. BRD4-NUT and p300 cooperate to induce hyperacetylated chromatin domains through a chromatin acetylation/binding loop feedforward mechanism. The mechanism schematized is based on data published by Reynoird and colleagues [3][1]. The scheme represents ouresearchers understanding of the mechanism underlying the initiation and propagation of hyperacetylated chromatin domains leading to the creation of multiple BRD4-NUT/p300-hyperacetylated chromatin foci. The initiation of the process corresponds to the binding of acetylated nucleosomes by BRD4, the recruitment of p300 by NUT and the enhancement of p300 catalytic activity, leading to the acetylation of adjacent nucleosomes. These new sites of histone acetylation recruit additional BRD4-NUT molecules, which in turn recruit p300 and a feedforward loop of histone acetylation-BRD4-NUT binding starts. The BRD4-NUT/p300 dependent histone acetylation propagation encounters opposing deacetylase activities constraining the acetylation propagation. The limits of the acetylated chromatin foci are dynamic and could move forward or backward depending on the opposing activities of acetylation and deacetylation.
2. NUT Is Specifically Expressed in Post-Meiotic Phases of Spermatogenesis
Spermatogenesis is a highly specialized differentiation program encompassing three characteristic stages, which include mitotic (or pre-meiotic), meiotic and post-meiotic phases. In the testis, a population of diploid adult stem cells, known as spermatogonia, is either maintained as stem cells or committed to differentiation following mitotic divisions. The committed spermatogonia-derived cells become spermatocytes while they undergo two meiotic divisions to generate haploid post-meiotic cells named spermatids.
The post-meiotic maturation of spermatids involves several major morphological and functional changes including a genome-wide chromatin remodelling and genome reorganization, resulting in the extreme compaction of the male genome in mature sperm cells [5,16,17][3][14][15]. Indeed, in these cells, the universal mode of eukaryotic chromatin organization, based on units named nucleosomes (each nucleosome consists of an octamer of 4 core histones, H2A, H2B, H3 and H4, around which the DNA is wrapped) shifts toward a new genome packaging structure based on the association of DNA with non-histone small basic proteins called protamines [5][3]. This histone-to-protamine transition allows a very tight compaction of the genome in mature spermatozoa, which is essential to protect the paternal genome during its transportation out of the parent organism through harsh environmental conditions in order to reach the female gamete, the oocyte [16][14].
The molecular basis of the essential process of histone-to-protamine replacement has remained one of the most obscure of all biological phenomena. The initial knowledge of the molecular basis of histone-to-protamine replacement was limited to only a few facts including a genome-wide hyperacetylation of histones [7][5] prior to their replacement and the expression of several highly specific histone variants by spermatogenic cells [17][15]. However, when this project was started in ouresearchers laboratory, nothing was known about the mechanisms driving this histone hyperacetylation, or its role in histone eviction, or the role of histone variants.
The early published work on the activity of the BRD4-NUT fusion protein [3][1] identified NUT as an excellent candidate in driving the histone hyperacetylation associated with histone eviction in spermatids, which prompted ouresearchers laboratory to specifically consider its role in this process during mouse spermatogenesis.
This hypothesis was reenforced by considering the pattern of NUT expression during spermatogenesis. Indeed, at the mRNA as well as protein levels, NUT first appears in post-meiotic cells, just at the time when histone hyperacetylation starts [4][2].
3. NUT Is Essential for Histone H4 Hyperacetylation and Histone-to-Protamine Replacement
To test ouresearchers hypothesis, Nut ko mice were generated. Nut ko male mice turned out to be infertile, with a total absence of spermatozoa. A more detailed analysis of spermatogenesis demonstrated that spermatids disappear at the time of protamine assembly and histone displacement [4][2]. This observation suggests that the absence of NUT could create an acute cell toxicity when cells prepare to set up the process of histone-to-protamine replacement.
Because of a possible role for NUT in histone hyperacetylation, an unbiased quantitative and qualitative proteomic analysis comparing the acetylation levels for each histone lysine position between wild-type spermatid cells expressing NUT and their Nut ko counterparts was carried out. Remarkably, this proteomic analysis, which was confirmed by immunoblotting, demonstrated that NUT is required for the acetylation of histone H4, specifically at K5 and K8. Since, as previously mentioned, the co-occurrence of H4K5acK8ac is known as a signature of hyperacetylated H4 and that it is required for the binding of BRDT itself involved in downstream events [10][8], these results supported the hypothesis that NUT is a critical actor in mediating the observed histone hyperacetylation in spermatids at the time of histone eviction [4][2].
4. Genetic Interaction between NUT and BRDT
An unbiased histone acetylome analysis identified H4 lysine 5 and lysine 8 (H4K5K8) as particularly sensitive to NUT-mediated H4 acetylation [4][2]. Considering that the previous structural studies of BRDT’s bromodomains demonstrated that BRDT’s BD1 precisely recognizes H4K5acK8ac [11][9], these new results appeared very relevant and exciting.
This observation suggested that BRDT’s action in spermatids would be dependent on the prior action of NUT, and therefore the prediction was that spermatids from Nut ko mice should show a phenotype similar as that of spermatids from Brdt delta-BD1 mice, expressing a BRDT mutant protein deleted for its first bromodomain (BRDT delta-BD1). Previous investigations had shown that spermatids expressing BRDT delta-BD1 are unable to replace their histones [10][8]. In these cells, although protamines are normally expressed, they are not incorporated and instead they accumulate around the nucleus [10,18][8][16]. Remarkably, exactly the same phenotype is observed in Nut ko spermatids. Indeed, these cells express protamines but, as observed in Brdt delta-BD1 spermatids, these protamines remain around the nucleus and the histones are not displaced [4][2].
Hence, these observations strongly support the hypothesis that NUT and BRDT function along the same molecular pathway, leading to histone-to-protamine replacement. The requirement of NUT to induce the acetylation of H4 on K5 and K8 and the ability of BRDT BD1 to specifically recognize and bind H4K5acK8ac and to displace histones, perfectly explain the observed genetic interaction between NUT and BRDT (Figure 2).
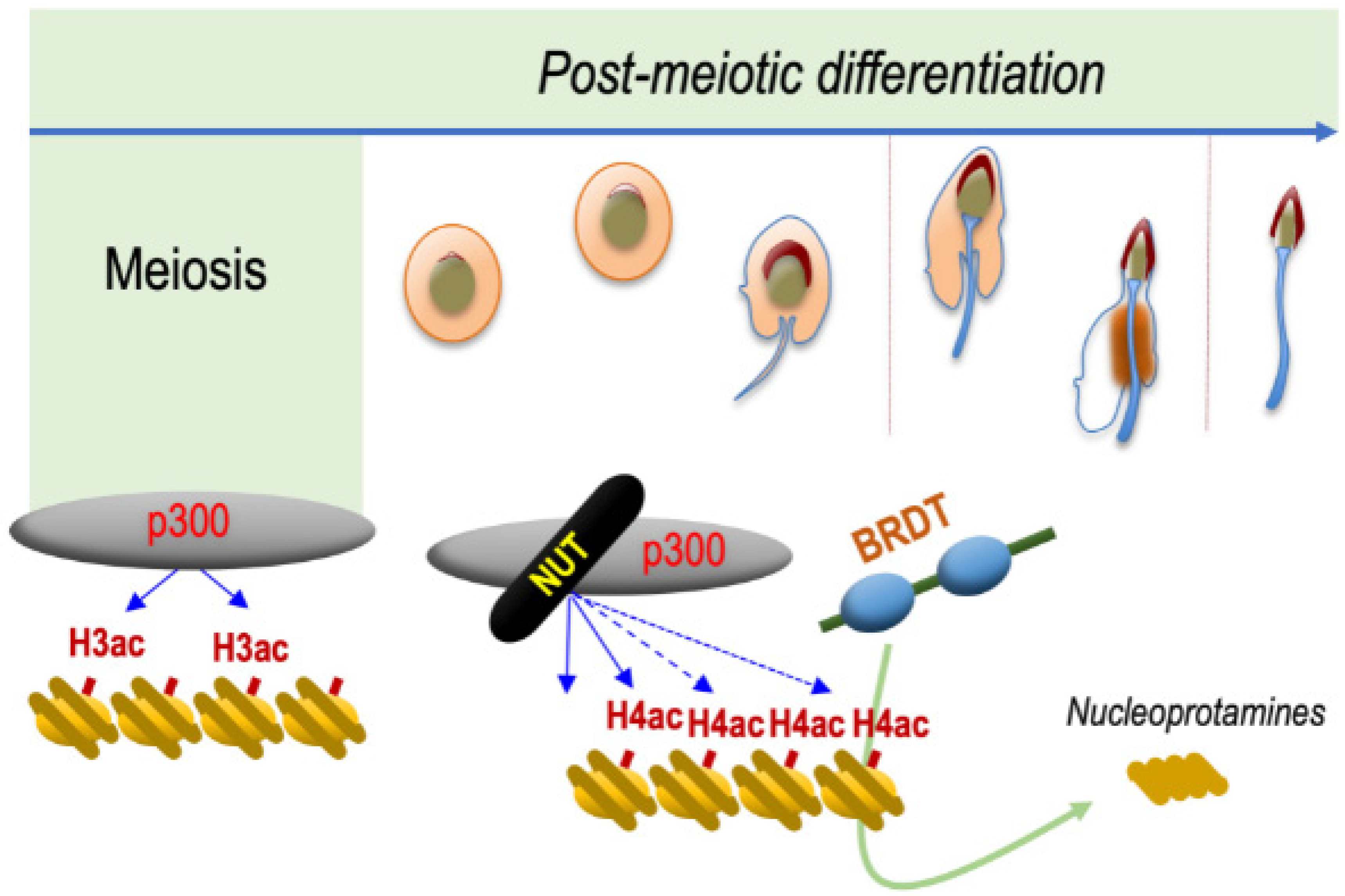
Figure 2. NUT is specifically expressed in post-meiotic spermatogenic cells and drives histone H4 hyperacetylation preceding histone eviction. The mechanism schematized is based on data published by Shiota and colleagues [4][2]. NUT, which is specifically expressed in spermatids, recruits CBP/p300, which are already expressed in all spermatogenic cell types. NUT stimulates their catalytic activity leading to the acetylation of H4 on both its lysines 5 and 8, which is required for the binding of the first bromodomain of BRDT. The binding of BRDT to H4K5acK8ac leads to the replacement of histones by protamines and the final compaction of the haploid male genome.
References
- Reynoird, N.; Schwartz, B.E.; Delvecchio, M.; Sadoul, K.; Meyers, D.; Mukherjee, C.; Caron, C.; Kimura, H.; Rousseaux, S.; Cole, P.A.; et al. Oncogenesis by Sequestration of CBP/P300 in Transcriptionally Inactive Hyperacetylated Chromatin Domains. EMBO J. 2010, 29, 2943–2952.
- Shiota, H.; Barral, S.; Buchou, T.; Tan, M.; Couté, Y.; Charbonnier, G.; Reynoird, N.; Boussouar, F.; Gérard, M.; Zhu, M.; et al. Nut Directs P300-Dependent, Genome-Wide H4 Hyperacetylation in Male Germ Cells. Cell Rep. 2018, 24, 3477–3487.e6.
- Bao, J.; Bedford, M.T. Epigenetic Regulation of the Histone-to-Protamine Transition during Spermiogenesis. Reproduction 2016, 151, 55–70.
- Hazzouri, M.; Pivot-Pajot, C.; Faure, A.K.; Usson, Y.; Pelletier, R.; Sèle, B.; Khochbin, S.; Rousseaux, S. Regulated Hyperacetylation of Core Histones during Mouse Spermatogenesis: Involvement of Histone Deacetylases. Eur. J. Cell Biol. 2000, 79, 950–960.
- Goudarzi, A.; Shiota, H.; Rousseaux, S.; Khochbin, S. Genome-Scale Acetylation-Dependent Histone Eviction during Spermatogenesis. J. Mol. Biol. 2014, 426, 3342–3349.
- Dhalluin, C.; Carlson, J.E.; Zeng, L.; He, C.; Aggarwal, A.K.; Zhou, M.M. Structure and Ligand of a Histone Acetyltransferase Bromodomain. Nature 1999, 399, 491–496.
- Pivot-Pajot, C.; Caron, C.; Govin, J.; Vion, A.; Rousseaux, S.; Khochbin, S. Acetylation-Dependent Chromatin Reorganization by BRDT, a Testis-Specific Bromodomain-Containing Protein. Mol. Cell Biol. 2003, 23, 5354–5365.
- Gaucher, J.; Boussouar, F.; Montellier, E.; Curtet, S.; Buchou, T.; Bertrand, S.; Hery, P.; Jounier, S.; Depaux, A.; Vitte, A.-L.; et al. Bromodomain-Dependent Stage-Specific Male Genome Programming by Brdt. EMBO J. 2012, 31, 3809–3820.
- Morinière, J.; Rousseaux, S.; Steuerwald, U.; Soler-López, M.; Curtet, S.; Vitte, A.-L.; Govin, J.; Gaucher, J.; Sadoul, K.; Hart, D.J.; et al. Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain. Nature 2009, 461, 664–668.
- Zhang, K.; Williams, K.E.; Huang, L.; Yau, P.; Siino, J.S.; Bradbury, E.M.; Jones, P.R.; Minch, M.J.; Burlingame, A.L. Histone Acetylation and Deacetylation: Identification of Acetylation and Methylation Sites of HeLa Histone H4 by Mass Spectrometry. Mol. Cell Proteom. 2002, 1, 500–508.
- Garcia, B.A.; Hake, S.B.; Diaz, R.L.; Kauer, M.; Morris, S.A.; Recht, J.; Shabanowitz, J.; Mishra, N.; Strahl, B.D.; Allis, C.D.; et al. Organismal Differences in Post-Translational Modifications in Histones H3 and H4. J. Biol. Chem. 2007, 282, 7641–7655.
- Sasaki, K.; Ito, T.; Nishino, N.; Khochbin, S.; Yoshida, M. Real-Time Imaging of Histone H4 Hyperacetylation in Living Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 16257–16262.
- Miller, T.C.R.; Simon, B.; Rybin, V.; Grötsch, H.; Curtet, S.; Khochbin, S.; Carlomagno, T.; Müller, C.W. A Bromodomain-DNA Interaction Facilitates Acetylation-Dependent Bivalent Nucleosome Recognition by the BET Protein BRDT. Nat. Commun. 2016, 7, 13855.
- Gaucher, J.; Reynoird, N.; Montellier, E.; Boussouar, F.; Rousseaux, S.; Khochbin, S. From Meiosis to Postmeiotic Events: The Secrets of Histone Disappearance. FEBS J. 2010, 277, 599–604.
- Hoghoughi, N.; Barral, S.; Vargas, A.; Rousseaux, S.; Khochbin, S. Histone Variants: Essential Actors in Male Genome Programming. J. Biochem. 2018, 163, 97–103.
- Rezaei-Gazik, M.; Vargas, A.; Amiri-Yekta, A.; Vitte, A.-L.; Akbari, A.; Barral, S.; Esmaeili, V.; Chuffart, F.; Sadighi-Gilani, M.A.; Couté, Y.; et al. Direct Visualization of Pre-Protamine 2 Detects Protamine Assembly Failures and Predicts ICSI Success. Mol. Hum. Reprod. 2022, 28, gaac004.
More
