Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Nora Tang and Version 1 by Bruno Silvester Lopes.
Antimicrobial Peptides (AMPs) can be divided into four major categories: those derived from mammals (human host defence peptides), amphibians and fish, microorganisms, and insects. The AMPs found in oceans have also attracted widespread attention.
- antimicrobial peptides
- biofilms
- Gram-negative bacteria
- WHO priority pathogens
1. Mammalian AMPs
Mammalian antimicrobial peptides are found in human, sheep, cattle, and other vertebrates. Cathelicidins and defensins are the two main families of Antimicrobial Peptides (AMPs) [24][1]. Cathelicidin are polypeptides that are primarily stored in the lysosomes of macrophages and polymorphonuclear leukocytes in humans and are encoded by the CAMP gene, which encodes the peptide precursor CAP-18, which is processed by proteinase 3-mediated extracellular cleavage into the active form LL-37 [63][2]. It is the only peptide of the cathelicidin family found in the human body [64][3].
The first reported AMP of animal origin was defensin, which was isolated from rabbit and guinea pig granulocytes in 1956 [65,66][4][5]. Defensins can be divided into α-, β-, and θ-defensins depending on the positions of disulfide bonds (Reddy et al., 2004). α-defensins are small peptides with 29–35 amino acid residues and three intramolecular disulfide linkages. Humans are known to possess six α-defensins, four human neutrophil peptides (HNP 1–4) expressed by granulocytes and certain lymphocytes, and two (HD-5 and 6) expressed by intestinal Paneth cells [35][6].
The human β-defensins are of four types (HBD 1–4) and are composed of up to 45 residues that are expressed by the epithelial skin cells, and psoriatic scales and the synthesis of these peptides can be induced by cytokines such as TNF- α and IL-1 β [36,37][7][8]. Human host defensins can protect humans from microbial infections but show different levels of expression at every stage of human growth. For example, cathelicidin LL-37, is usually detected in the skin of new-borns, whereas human beta-defensin 2 (HBD-2) is often expressed in the elderly [67][9]. Human defensin peptides are present in several parts of the body, including the skin, eyes, ears, mouth, respiratory tract, lung, intestine, urethra, tracheal epithelium, and at elevated levels (HBD-2 in particular) in human keratinocytes exposed to pathogenic bacteria [24,68][1][10]. AMPs from human breast milk can play an important role in decreasing the morbidity and mortality of breast-feeding infants [69][11]. A recent study showed that the levels of AMPs were similar in the breast-milk-fed preterm infants with and without late-onset neonatal sepsis, but since preterm infants with late-onset neonatal sepsis consumed significantly less breast milk, the levels of milk AMPs were low. This indicates that the concentrations of lactoferrin and defensins in preterm breast milk have antimicrobial activity against neonatal pathogens [69][11]. Dairy is an important source of AMPs, which are produced by the enzymatic hydrolysis of milk, of which lactoferricin B (LfcinB) is well-known for its antimicrobial properties that affect intracellular activities, leading to bacterial inhibition [38][12]. In addition to antimicrobial activity, cathelicidins and defensins have also been shown to affect immune regulation, apoptosis, and wound healing [39][13]. The mechanisms of action of antimicrobial peptides are summarised in Figure 21.
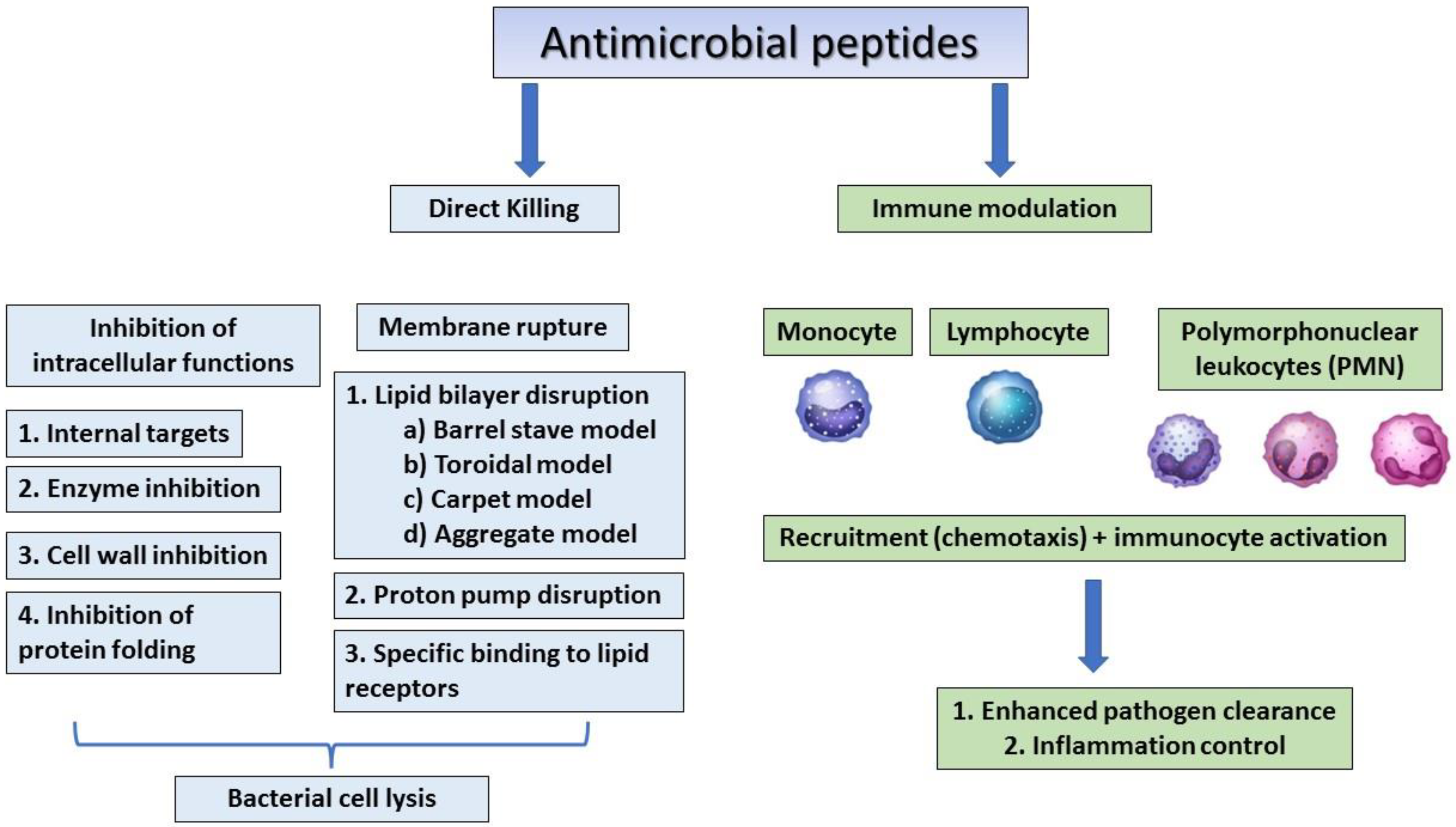
Figure 21.
The general mechanism of action of antimicrobial peptides.
2. Amphibian and Fish AMPs
Antimicrobial peptides from amphibians play an important role in the protection of amphibian species from pathogens and global decline [70,71][14][15]. Frogs and toads are considered the main and the largest source of amphibian AMPs. Cancrin was the first AMP to be discovered from the sea amphibian Rana cancrivora, a crab-eating frog [72][16]. The most important being the magainins, which are peptides constitutionally produced by mucous glands in the skin of several frogs of genera such as Xenopus, Silurana, Hymenochirus, and Pseudhymenochirus possessing antibacterial, antifungal, and antitumor properties. Others include peptides such as bombinins, cathelicidin, maximins, phylloxin, plasticins, and palustrin, which are antibacterial; buforin, esculentins, and pseudins, which are antibacterial and antifungal; dermaseptins that are antiviral, antibacterial, antifungal, and anticancer; fallaxin, which is antibacterial; and leishmanicidal and ranateurins, which show antibacterial and anticancer properties [40][17]. Cathelicidins, defensins, hepicidins, piscidins, and histone-derived peptides are AMPs found in fish and also occur in humans, cattle, pigs, and horses [73][18]. The broad-spectrum antimicrobial peptides (HFIAP-1, -2, and -3) were the first cathelicidins that were isolated from the intestinal tissues of Myxine glutinosa (Atlantic hagfish), which possessed broad-spectrum antimicrobial activity against a number of Gram-positive and -negative bacteria [41][19]. The cathelicidin codCATH, isolated from Atlantic Cod (Gadus morhua), which is rich in glycine residues, shows activity against Gram-negative bacteria, whereas rtCATH1 (R146-P181) and rtCATH2 (R143-I178), isolated from Rainbow Trout, are active against Lactococcus garvieae and other Gram-negative fish pathogens [42,43][20][21]. Fish defensins/defensin-like proteins consisting of six conserved cysteine motifs were initially discovered in zebrafish, Fugu, and tetraodon using a database-mining approach [74][22]. These show potent activity against bacteria and fish-specific viruses, e.g., cod defensin defb, identified in Atlantic cod, shows antibacterial activity against the Gram-positive bacteria Planococcus citreus and Micrococcus luteus [44][23], whereas EcDefensin, isolated from the orange-spotted grouper, Epinephelus coioides, is shown to inhibit the replication of a DNA virus, Singapore Grouper Iridovirus (SGIV), and an RNA marine fish virus, viral nervous necrosis virus (VNNV) [75][24]. Fish hepcidins are rich in cysteine and are hormones involved in iron regulation that share a β-sheet-composed hairpin structure linked via four disulfide bonds, e.g., Om-hep1 from Oryzias melastigmus is active against Gram-positive bacteria such as C. glutamicum and S. aureus and Gram-negative pathogens such as E. coli MC1061, A. hydrophila, and Pseudomonas stutzeri [76][25]. Piscidins share an α-helical structure similar to magainins and are linear AMPs, which are amphipathic in nature [77][26]. Pleurocidin is a magainin isolated from winter flounder (Pleuronectes americanus) and is active against both Gram-positive and -negative pathogens [45][27].
3. AMPs from Microorganisms
3.1. AMPs from Gram-Positive Bacteria
Antimicrobial peptides such as nisin and gramicidin are produced by Gram-positive bacteria such as Lactococcus lactis and Bacillus brevis [78][28]. Both ribosomally and non-ribosomally synthesised AMPs have been found in Gram-positive bacteria [46][29].
Ribosomally synthesised bacterial AMPs are termed as bacteriocins [79][30]. There are four classes of bacteriocins produced by Gram-positive bacteria: (I) lantibiotics, (II) non-lantiboitics, (III) large-sized bacteriocins, and (IV) uniquely structured bacteriocins [78][28]. Small peptides (<5KDa, 19–38 amino acids), which are post-translationally modified and contain unusual amino acids, belong to the class of lantibiotics. Subclass 1a is positively charged and acts by forming pores (e.g., nisin, epidermin, gallidermin, and Pep5). Subclass 1b is negatively charged, globular, inflexible, and acts by inhibiting crucial enzymes in targeted pathogens (e.g., mersacidin, actagardine, and cinnamycin) [80][31]. Non-lantibiotics are small, heat-stable peptides with limited post-translational modification. They act by increasing cell permeability by pore formation. Subclass IIa are disulfide-containing linear AMPs (e.g., pediocin PA-1 and enterocin P) [81[32][33],82], subclass IIb contain α and β subunits (e.g., plantaricin EF, lactococcin G, thermophilin 13, and lactacin F) [83][34], subclass IIc are small cyclic peptides with covalently linked N and C-termini (e.g., enterocin AS-48, gassericin A, circularin A, and lactocyclicin Q) [84][35], and subclass IId remain to be characterised (e.g., lactococcin A, B, and 972 and enterocinL50) [83][34]. Large-sized bacteriocins (>30 kDa) are also called bacteriolysins and are heat-labile peptides (e.g., zoocin A, lysostaphin, and enterolysin A) [85][36]. Uniquely structured bacteriocins contain amino acids, lipids, or carbohydrates and are susceptible to lipolytic or glycolytic enzymes (e.g., plantaricin S, leuconocin S, and lactocin 27) [86,87][37][38].
Non-ribosomally synthesised AMPs are cyclic heptapeptides consisting of a tripeptide side chain linked to an N-terminal fatty acyl tail and are obtained from the Gram-positive spore-forming soil bacterium Paenibacillus polymyxa [47,48][39][40]. So far, at least 10 different groups of polymyxin lipopeptides have been identified; they are polymyxins A, B, C, D, E, F, M, P, S, and T, with polymyxin B and E (colistin) being used clinically, and are active against critically important pathogens listed by the WHO such as A. baumannii, P. aeruginosa, and S. maltophilia [48][40]. Tridecaptins (A, B, and C) are a re-emerging class of non-ribosomal antibacterial peptides (NRAPs) that are isolated from Paenibacillus polymyxa AR-110, B-2, and E-23, respectively, and show potent activity against drug-resistant strains of Gram-negative pathogens, where they selectively bind to the Gram-negative analogue of peptidoglycan precursor lipid II on the outer leaflet of the inner membrane, disrupting the proton motive force [88][41].
3.2. AMPs from Gram-Negative Bacteria
Bacteriocins are AMPs that are isolated from Gram-negative bacteria such as E. coli, Klebsiella, and Pseudomonas spp. [49][42] that show a narrow-spectrum activity against Gram-negative pathogens. They are classified into four different classes: colicins, colicin-like bacteriocins, microcins, and phage tail-like bacteriocins.
Colicins (>10 kDa) are plasmid-encoded AMPs produced by E. coli and specifically bind to cell surface receptors before translocation through the outer membrane, periplasm, and inner membrane into the cell cytoplasm [50][43]. Colicin A, K, and U bind to BtuB, Tsx, and OmpA receptors, respectively [89][44]. Colicins are divided into three classes according to their mechanisms of action: (i) forming channels or pores in the cytoplasmic membrane (e.g., colicin A, B, and E1), (ii) DNA degrading (e.g., colicin E2, E7, E8, and E9), targeting rRNA (e.g., ColE3, ColE4, ColE6, and DF13) or tRNA (e.g., ColE5, ColE6, and Col D), and (iii) inhibition of murein and lipopolysaccharide synthesis (e.g., colicin M) [51,52][45][46].
Colicin-like bacteriocins are produced by Gram-negative bacteria such as P. aeruginosa (e.g., S-type pyocins) and Klebsiella spp. (klebicins) and are structurally and functionally similar to E. coli colicins [90,91][47][48]. S-type pyocins (AP41, S1–S5) are sensitive to proteases and induce cell death by cleaving DNA (AP41 and S1–S3) or RNA (S4) or via pore formation (S5) [92][49]. The action of klebicins occurs via endonuclease activity, pore formation, and/or by the degradation of peptidoglycan [91][48].
Microcins are small peptides (<10 kDa) produced by bacteria belonging to Enterobacteriaceae and are classified into two subclasses (e.g., Colicin V and Microcin C7) [50][43]. Subclass I (<5 kDa) undergo extensive post-translational modification compared to subclass II (>5–10 kDa), which are unmodified or slightly modified post-translationally [40][17].
High molecular weight cylindrical peptides with high similarity to the phage tail structure are referred to as phage tail-like bacteriocins [40][17]. These are also divided into two subclasses. Subclass I consists of R-type bacteriocin related to the contractile tail of phages belonging to the family Myoviridae. Subclass II consists of F-type bacteriocins related to Siphoviridae phage tails. R- and F-type pyocins are produced by Pseudomonas aeruginosa and are encoded in a gene cluster comprising a DNA region greater than 40 kb [50][43].
3.3. Fungal AMPs
Many fungal AMPs not only show inhibitory activity against common pathogenic fungi, such as Aspergillus and Candida spp. in humans, but also against yeast and filamentous fungi (e.g., Aspergillus flavus), which affect food and agriculture [24][1]. Fungal AMPs are divided into peptaibols (found in Trichoderma spp.) and defensins (found in Pseudoplectania, Coprinopsis, and Microsporum spp.) [40][17]. The fungal AMPs contain 5–21 amino acids, with a high proportion of non-proteinogenic amino acids, such as α-aminoisobutyric acid, and typically have an acylated N-terminal residue and an amino alcohol, such as phenylalaninol or leucenol, attached to the C-terminal [93][50]. Alamethicin is the most widely studied peptaibol isolated from T. viridea, which is active against both Gram-positive bacteria such as E. faecalis, S. hemolyticus, and S. aureus and Gram-negative bacteria such as E. coli, K. pneumoniae, and P. aeruginosa, with antimicrobial activity against fungi as well [93,94,95][50][51][52]. Peptaibols can be classified as short-chain, consisting of 5–10 amino acids; medium-chain, with 11–16 amino acids; and long-chain, with 17–21 amino acids, with the primary mechanism of action involving membrane disruption [53]. Peptaibols of ≥15 amino acids can form helical structures that oligomerise, forming ion channels in the membrane [96][54]. Peptaibols of <15 amino acids act by a combination of membrane disruption (e.g., the formation of transmembrane channels or by a barrel-stave mechanism) and also affect different molecular targets.
The fungal defensin-like peptides are cysteine-rich peptides that show high sequence and structural similarities to defensins from microorganisms, plants, and animals [74,97][22][55]. Plectasin isolated from Pseudoplectania nigrella was the first fungal defensin to be characterised, which showed inhibitory activity against Gram-positive bacteria such as S. pyogenes, C. diphtheriae, and S. aureus [54][56]. Micasin from Microsporum canis was found to show broad-spectrum antibacterial activity against both P. aeruginosa and methicillin-resistant S. aureus by affecting protein folding [55][57].
3.4. Viral and Bacteriophage AMPs
Antiviral drug resistance is an increasing concern in immunocompromised patients, with the limited efficiency of commonly used antiviral drugs making viral AMPs ideal candidates as potential therapeutic agents [98][58]. Antiviral agents act at different levels, such as inhibiting the activity of viral reverse transcriptase, retroviral integrase, or proteases or the inhibition of the transport of circular viral DNA to the nucleus, impairing cellular processes [99][59]. Antiviral AMPs integrate into viral envelopes, causing membrane instability and thereby preventing the viruses from infecting host cells [100][60]. Melittin (the main compound found in the venom of the European honeybee Apis mellifera), which also has anticancer activity, has an inhibitory activity against both enveloped and non-enveloped viruses, including coxsackievirus, enterovirus, influenza A viruses, human immunodeficiency virus (HIV), herpes simplex virus (HSV), Junín virus (JV), respiratory syncytial virus (RSV), vesicular stomatitis virus (VSV), and tobacco mosaic virus (TMV) [56][61]. It has also been suggested that melittin impedes cell fusion in HSV-1 glycoprotein K mutants by interfering with the activity of sodium potassium ATPase, an essential enzyme involved in the membrane fusion process [101][62]. Antiviral AMPs can also prevent viral particles from entering the host cells by binding to specific mammalian cell receptors, e.g., lactoferrin inhibits HSV infections by binding to heparan sulfate proteoglycans, which play an important role in the attachment of HSV viral particles to the host cell surface, thereby blocking any interactions with the virus receptor [102][63].
Bacteriophages are a type of viruses that infect bacteria, and the word “bacteriophage” literally refers to “bacteria eating”. These viruses multiply in the presence of bacteria and kill them to release their progeny. This mechanism is used in clinical treatment to kill the target bacteria, and many studies are ongoing to develop phages into a more potent alternative. In vitro and in vivo works have shown that phages have a high efficacy in killing multidrug-resistant bacteria [103][64]. Whole phage-based treatments are time-consuming; thus, phage-derived antimicrobial peptides such as endolysins are being studied as an alternative medicine to treat bacterial infections.
Many phage proteins, including endolysins, virion-associated peptidoglycan hydrolases (VAPGHs), depolymerases, and holins, display antibacterial activity [104,105][65][66]. Phage AMPs are of two types, phage-encoded bacteriolytic proteins produced at the end of the lytic cycle and phage-tail complexes [104,106][65][67]. Based on the enzymatic activity, endolysins can be divided into four major types, namely acetylmuramidases, glucosaminidases, amidases, transglycosylases, and endopeptidases [107][68]. Each endolysin is known to have a specific enzymatic function. The endolysin structure dictates the function and activity of the endolysin. In general, endolysin is made up of two active sites, one at the N-terminal linked by a short flanking region to the cell-wall-binding domain at the C-terminal [108][69]. The endolysin specific to the Gram-positive bacteria are known to resemble fungal cellulases, and Gram-negative-specific endolysins have multiple globular structures that are drawn from the primary endolysin motifs [109][70].
Phage lysins are peptidoglycan-hydrolysing enzymes ranging from 25 to 40 kDa. They act by weakening the peptidoglycan bacterial cell wall, creating holes that permit phage progeny to exit the cell, resulting in rapid bactericidal activity. They may also possess synergistic activity with cell-wall-inhibiting antibiotics, antibiofilm activity, and are heat-stable up to a temperature of 50 °C [57,58][71][72]. Examples include LysAB2 P3, which shows inhibitory activity against Acinetobacter baumannii [110][73], and PlyV12, a phage lysin that exhibits broad bactericidal activity against enterococci and other Gram-positive pathogens such as S. pyogenes and S. aureus [111][74]. VAPGHs are genus/species-specific lytic enzymes that are encoded by double-stranded DNA phages that specifically degrade peptidoglycan. They have a C-terminal cell-wall-binding domain and one or more N-terminal catalytic domains [112][75]. VAPGHs can be classified into three categories based on the peptidoglycan cleavage site, namely, glycosidases (muramidases that cleave between N-acetylmuramic acid and N-acetylglucosamine residues, similar to lysozyme), amidases (those that cleave between N-acetylmuramic acid and the first highly conserved L-alanine residue), and endopeptidases (cleave between two amino acid residues) [113][76]. VAPGHs are active against both Gram-positive and -negative bacteria. VAPGH HydH5 of Φ vB_SauS-phiIPLA88 is active against S. aureus, whereas Phage Φ6 contains protein P5, an endopeptidase that shows inhibitory activity against P. aeruginosa [112,114][75][77]. Phage polysaccharide depolymerases are enzymes that degrade the macromolecule carbohydrates of the bacterial cell wall [115][78]. Klebsiella phage ΦK64-1 encodes several depolymerases that show activity against a number of Klebsiella capsular polysaccharides, whereas P. putida phage AF degrades the extracellular polysaccharides involved in the formation of the biofilm matrix of P. putida [116,117,118][79][80][81]. Hence, depolymerases may be effective against bacteria producing biofilm such as P. mirabilis, E. coli, S. suis, K. pneumoniae, and P. aeruginosa and in the treatment of biofilms formed by foodborne bacteria, such as Campylobacter and Salmonella [119,120,121,122,123][82][83][84][85][86]. OmniLytics Inc. (Sandy, UT, USA), has developed two products, BacWashTM for Salmonella and FinalyseTM for E. coli O157:H7, marketed by Elanco (Greenfield, IN, USA). Intralytix Inc. (Baltimore, MD, USA) developed three phage products, ListShieldTM, EcoShieldTM, and SalmoFreshTM, to be used in the food industry against L. monocytogenes, E. coli, and Salmonella, respectively, which can help in making food more secure, thereby protecting public health [123][86]. Holins are a diverse group of small hydrophobic proteins produced by dsDNA bacteriophages (<150 amino acids) that are involved in the regulation of bacterial lysis time by guiding the phage muramidases to the peptidoglycan layer [59][87]. Canonical holins form large pores on one side of the bacteria, locally exposing the peptidoglycans to cytoplasmic canonical endolysin molecules [124][88]. Pinholins form small pores that result in membrane depolarisation, triggering signal-arrest-release (SAR) endolysin activation and the degradation of peptidoglycans in the whole cellular periplasmic space [124][88]. The S. aureus bacteriophage GH15 produces the holin HolGH15, which has broad antibacterial activity against the foodborne pathogen L. monocytogenes [60][89]. Phage-derived endolysins are a powerful weapon for combatting antibiotic resistance, as they exhibit rapid activity against bacterial cells, and their remarkable ability to lyse bacterial cells, even in the absence of bacterial multiplication, makes them a superior medicine over whole phage therapy.
3.5. Insect AMPs
Antimicrobial peptides are mainly synthesized in the fat bodies and blood cells of insects and are a source of strong adaptability and survival [125][90]. Insect AMPs exhibit an antimicrobial effect by disrupting the cell membrane and prevent microbes from developing drug resistance [126][91]. Cecropin A, found in cecropia moth (Hyalophora cecropia) and bees, shows activity against different inflammatory diseases and cancers [61][92]. Insect AMPs such as cecropins, ponericins, defensins, lebocins, drosocin, metchnikowin, gloverins, diptericins, and attacins are a heterogeneous group of immunity-related proteins that exhibit an antimicrobial effect, mainly against Gram-negative bacteria [127][93]. A peptide derived from the royal jelly of honeybees, Jellein, shows promising effects on several bacteria and fungi, and its conjugated form with lauric acid can inhibit the parasite Leishmania major [62][94].
References
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779.
- Wang, G. Structures of Human Host Defense Cathelicidin LL-37 and its Smallest Antimicrobial Peptide KR-12 in Lipid Micelles. J. Biol. Chem. 2008, 283, 32637–32643.
- Dürr, U.H.; Sudheendra, U.; Ramamoorthy, A. LL-37, the Only Human Member of the Cathelicidin Family of Antimicrobial Peptides. Biochim. Biophys. Acta (BBA)—Biomembr. 2006, 1758, 1408–1425.
- Hirsch, J.G. Phagocytin: A Bactericidal Substance from Polymorphonuclear Leucocytes. J. Exp. Med. 1956, 103, 589–611.
- Zeya, H.I.; Spitznagel, J.K. Cationic Proteins of Polymorphonuclear Leukocyte Lysosomes. I. Resolution of Antibacterial and Enzymatic Activities. J. Bacteriol. 1966, 91, 750–754.
- Ouellette, A.J.; Bevins, C.L. Paneth Cell Defensins and Innate Immunity of the Small Bowel. Inflamm. Bowel Dis. 2001, 7, 43–50.
- Hoover, D.M.; Chertov, O.; Lubkowski, J. The Structure of Human Β-Defensin-1: New Insights into Structural Properties of Β-Defensins. J. Biol. Chem. 2001, 276, 39021–39026.
- Schröder, J.; Harder, J. Human Beta-Defensin-2. Int. J. Biochem. Cell Biol. 1999, 31, 645–651.
- Gschwandtner, M.; Zhong, S.; Tschachler, A.; Mlitz, V.; Karner, S.; Elbe-Bürger, A.; Mildner, M. Fetal Human Keratinocytes Produce Large Amounts of Antimicrobial Peptides: Involvement of Histone-Methylation Processes. J. Investig. Dermatol. 2014, 134, 2192–2201.
- Harder, J.; Bartels, J.; Christophers, E.; Schröder, J. A Peptide Antibiotic from Human Skin. Nature 1997, 387, 861.
- Trend, S.; Strunk, T.; Hibbert, J.; Kok, C.H.; Zhang, G.; Doherty, D.A.; Richmond, P.; Burgner, D.; Simmer, K.; Davidson, D.J. Antimicrobial Protein and Peptide Concentrations and Activity in Human Breast Milk Consumed by Preterm Infants at Risk of Late-Onset Neonatal Sepsis. PLoS ONE 2015, 10, e0117038.
- Tu, Y.; Ho, Y.; Chuang, Y.; Chen, P.; Chen, C. Identification of Lactoferricin B Intracellular Targets using an Escherichia coli Proteome Chip. PLoS ONE 2011, 6, e28197.
- Wang, G. Human Antimicrobial Peptides and Proteins. Pharmaceuticals 2014, 7, 545–594.
- Rollins-Smith, L.A. The Role of Amphibian Antimicrobial Peptides in Protection of Amphibians from Pathogens Linked to Global Amphibian Declines. Biochim. Biophys. Acta (BBA)—Biomembr. 2009, 1788, 1593–1599.
- Varga, J.F.; Bui-Marinos, M.P.; Katzenback, B.A. Frog Skin Innate Immune Defences: Sensing and Surviving Pathogens. Front. Immunol. 2019, 9, 3128.
- Lu, Y.; Ma, Y.; Wang, X.; Liang, J.; Zhang, C.; Zhang, K.; Lin, G.; Lai, R. The First Antimicrobial Peptide from Sea Amphibian. Mol. Immunol. 2008, 45, 678–681.
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691.
- Masso-Silva, J.A.; Diamond, G. Antimicrobial Peptides from Fish. Pharmaceuticals 2014, 7, 265–310.
- Basañez, G.; Shinnar, A.E.; Zimmerberg, J. Interaction of Hagfish Cathelicidin Antimicrobial Peptides with Model Lipid Membranes. FEBS Lett. 2002, 532, 115–120.
- Broekman, D.C.; Zenz, A.; Gudmundsdottir, B.K.; Lohner, K.; Maier, V.H.; Gudmundsson, G.H. Functional Characterization of codCath, the Mature Cathelicidin Antimicrobial Peptide from Atlantic Cod (Gadus Morhua). Peptides 2011, 32, 2044–2051.
- Chang, C.; Zhang, Y.; Zou, J.; Nie, P.; Secombes, C.J. Two Cathelicidin Genes are Present in both Rainbow Trout (Oncorhynchus mykiss) and Atlantic Salmon (Salmo salar). Antimicrob. Agents Chemother. 2006, 50, 185–195.
- Zou, J.; Mercier, C.; Koussounadis, A.; Secombes, C. Discovery of Multiple Beta-Defensin Like Homologues in Teleost Fish. Mol. Immunol. 2007, 44, 638–647.
- Ruangsri, J.; Kitani, Y.; Kiron, V.; Lokesh, J.; Brinchmann, M.F.; Karlsen, B.O.; Fernandes, J.M. A Novel Beta-Defensin Antimicrobial Peptide in Atlantic Cod with Stimulatory Effect on Phagocytic Activity. PLoS ONE 2013, 8, e62302.
- Guo, M.; Wei, J.; Huang, X.; Huang, Y.; Qin, Q. Antiviral Effects of Β-Defensin Derived from Orange-Spotted Grouper (Epinephelus coioides). Fish Shellfish. Immunol. 2012, 32, 828–838.
- Cai, L.; Cai, J.; Liu, H.; Fan, D.; Peng, H.; Wang, K. Recombinant Medaka (Oryzias melastigmus) Pro-Hepcidin: Multifunctional Characterization. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 161, 140–147.
- Fernandes, J.M.; Ruangsri, J.; Kiron, V. Atlantic Cod Piscidin and its Diversification through Positive Selection. PLoS ONE 2010, 5, e9501.
- Cole, A.M.; Weis, P.; Diamond, G. Isolation and Characterization of Pleurocidin, an Antimicrobial Peptide in the Skin Secretions of Winter Flounder. J. Biol. Chem. 1997, 272, 12008–12013.
- Cao, J.; de la Fuente-Nunez, C.; Ou, R.W.; Torres, M.D.T.; Pande, S.G.; Sinskey, A.J.; Lu, T.K. Yeast-Based Synthetic Biology Platform for Antimicrobial Peptide Production. ACS Synth. Biol. 2018, 7, 896–902.
- Diep, D.B.; Nes, I.F. Ribosomally Synthesized Antibacterial Peptides in Gram Positive Bacteria. Curr. Drug Targets 2002, 3, 107–122.
- Yeaman, M.R.; Yount, N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55.
- Brötz, H.; Sahl, H. New Insights into the Mechanism of Action of Lantibiotics—Diverse Biological Effects by Binding to the Same Molecular Target. J. Antimicrob. Chemother. 2000, 46, 1–6.
- Devi, S.M.; Halami, P.M. Detection and Characterization of Pediocin PA-1/AcH Like Bacteriocin Producing Lactic Acid Bacteria. Curr. Microbiol. 2011, 63, 181–185.
- Cintas, L.M.; Casaus, P.; Havarstein, L.S.; Hernandez, P.E.; Nes, I.F. Biochemical and Genetic Characterization of Enterocin P, a Novel Sec-Dependent Bacteriocin from Enterococcus faecium P13 with a Broad Antimicrobial Spectrum. Appl. Environ. Microbiol. 1997, 63, 4321–4330.
- Nissen-Meyer, J.; Oppegård, C.; Rogne, P.; Haugen, H.S.; Kristiansen, P.E. Structure and Mode-of-Action of the Two-Peptide (Class-IIb) Bacteriocins. Probiotics Antimicrob. Proteins 2010, 2, 52–60.
- Rea, M.C.; Ross, R.P.; Cotter, P.D.; Hill, C. Classification of Bacteriocins from Gram-Positive Bacteria. In Prokaryotic Antimicrobial Peptides; Springer: New York, NY, USA, 2011; pp. 29–53.
- Heng, N.C.; Tagg, J.R. What’s in a Name? Class Distinction for Bacteriocins. Nat. Rev. Microbiol. 2006, 4, 160.
- Da Silva Sabo, S.; Vitolo, M.; González, J.M.D.; de Souza Oliveira, R.P. Overview of Lactobacillus Plantarum as a Promising Bacteriocin Producer among Lactic Acid Bacteria. Food Res. Int. 2014, 64, 527–536.
- De Martinis, E.; Alves, V.; Franco, B. Fundamentals and Perspectives for the use of Bacteriocins Produced by Lactic Acid Bacteria in Meat Products. Food Rev. Int. 2002, 18, 191–208.
- Stansly, P.G.; Shepherd, R.G.; White, H.J. Polymyxin: A New Chemotherapeutic Agent. Bull. Johns Hopkins Hosp. 1947, 81, 43–54.
- Velkov, T.; Thompson, P.E.; Azad, M.A.; Roberts, K.D.; Bergen, P.J. History, Chemistry and Antibacterial Spectrum. In Polymyxin Antibiotics: From Laboratory Bench to Bedside; Springer: Cham, Switzerland, 2019; pp. 15–36.
- Bann, S.J.; Ballantine, R.D.; Cochrane, S.A. The Tridecaptins: Non-Ribosomal Peptides that Selectively Target Gram-Negative Bacteria. RSC Med. Chem. 2021, 12, 538–551.
- Riley, M.A.; Chavan, M.A. Bacteriocins; Springer: Cham, Switzerland, 2007.
- Johnstone, B.A.; Christie, M.P.; Morton, C.J.; Parker, M.W. X-ray crystallography shines a light on pore-forming toxins. Methods Enzymol. 2021, 649, 1–46.
- Cascales, E.; Buchanan, S.K.; Duché, D.; Kleanthous, C.; Lloubes, R.; Postle, K.; Riley, M.; Slatin, S.; Cavard, D. Colicin Biology. Microbiol. Mol. Biol. Rev. 2007, 71, 158–229.
- Gillor, O.; Kirkup, B.C.; Riley, M.A. Colicins and Microcins: The Next Generation Antimicrobials. Adv. Appl. Microbiol. 2004, 54, 129–146.
- Papadakos, G.; Wojdyla, J.A.; Kleanthous, C. Nuclease Colicins and their Immunity Proteins. Q. Rev. Biophys. 2012, 45, 57–103.
- Parret, A.H.; De Mot, R. Bacteria Killing their Own Kind: Novel Bacteriocins of Pseudomonas and Other Γ-Proteobacteria. Trends Microbiol. 2002, 10, 107–112.
- Denkovskienė, E.; Paškevičius, Š.; Misiūnas, A.; Stočkūnaitė, B.; Starkevič, U.; Vitkauskienė, A.; Hahn-Löbmann, S.; Schulz, S.; Giritch, A.; Gleba, Y. Broad and Efficient Control of Klebsiella Pathogens by Peptidoglycan-Degrading and Pore-Forming Bacteriocins Klebicins. Sci. Rep. 2019, 9, 15422.
- Michel-Briand, Y.; Baysse, C. The Pyocins of Pseudomonas aeruginosa. Biochimie 2002, 84, 499–510.
- Bissett, J.; Gams, W.; Jaklitsch, W.; Samuels, G.J. Accepted Trichoderma Names in the Year 2015. IMA Fungus 2015, 6, 263–295.
- Meyer, C.; Reusser, F. A Polypeptide Antibacterial Agent Isolated from Trichoderma viride. Experientia 1967, 23, 85–86.
- Chugh, J.; Wallace, B. Peptaibols: Models for Ion Channels. Biochem. Soc. Trans. 2001, 29, 565–570.
- Ramachander Turaga, V. Peptaibols: Antimicrobial Peptides from Fungi. In Bioactive Natural Products in Drug Discovery; Springer: Singapore, 2020; pp. 713–730.
- Chugh, J.; Brückner, H.; Wallace, B. Model for a Helical Bundle Channel Based on the High-Resolution Crystal Structure of trichotoxin_A50E. Biochemistry 2002, 41, 12934–12941.
- Grishin, D.; Sokolov, N. Defensins are Natural Peptide Antibiotics of Higher Eukaryotes. Biochem. Suppl. Ser. B Biomed. Chem. 2014, 8, 11–18.
- Schneider, T.; Kruse, T.; Wimmer, R.; Wiedemann, I.; Sass, V.; Pag, U.; Jansen, A.; Nielsen, A.K.; Mygind, P.H.; Raventós, D.S. Plectasin, a Fungal Defensin, Targets the Bacterial Cell Wall Precursor Lipid II. Science 2010, 328, 1168–1172.
- Zhu, S.; Gao, B.; Harvey, P.J.; Craik, D.J. Dermatophytic Defensin with Antiinfective Potential. Proc. Natl. Acad. Sci. USA 2012, 109, 8495–8500.
- Nyanguile, O. Peptide Antiviral Strategies as an Alternative to Treat Lower Respiratory Viral Infections. Front. Immunol. 2019, 10, 1366.
- Goodsell, D.S. Illustrations of the HIV Life Cycle. In The Future of HIV-1 Therapeutics; Springer: Cham, Switzerland, 2015; pp. 243–252.
- Mulder, K.; Lima, L.A.; Miranda, V.; Dias, S.C.; Franco, O.L. Current Scenario of Peptide-Based Drugs: The Key Roles of Cationic Antitumor and Antiviral Peptides. Front. Microbiol. 2013, 4, 321.
- Memariani, H.; Memariani, M.; Moravvej, H.; Shahidi-Dadras, M. Melittin: A Venom-Derived Peptide with Promising Anti-Viral Properties. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 5–17.
- Matanic, V.C.A.; Castilla, V. Antiviral Activity of Antimicrobial Cationic Peptides against Junin Virus and Herpes Simplex Virus. Int. J. Antimicrob. Agents 2004, 23, 382–389.
- Andersen, J.H.; Jenssen, H.; Sandvik, K.; Gutteberg, T.J. Anti-HSV Activity of Lactoferrin and Lactoferricin Is Dependent on the Presence of Heparan Sulphate at the Cell Surface. J. Med. Virol. 2004, 74, 262–271.
- Manohar, P.; Loh, B.; Namasivayam, E.; Loganathan, A.; Nachimuthu, R.; Leptihn, S. A Multiwell-Plate Caenorhabditis elegans Assay for Assessing the Therapeutic Potential of Bacteriophages against Clinical Pathogens. Microbiol. Spectr. 2022, 23, e0139321.
- Mirski, T.; Mizak, L.; Nakonieczna, A.; Gryko, R. Bacteriophages, Phage Endolysins and Antimicrobial Peptides-the Possibilities for their Common use to Combat Infections and in the Design of New Drugs. Ann. Agric. Environ. Med. 2019, 26, 203–209.
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage Endolysins as Novel Antimicrobials. Future Microbiol. 2012, 7, 1147–1171.
- Parisien, A.; Allain, B.; Zhang, J.; Mandeville, R.; Lan, C. Novel Alternatives to Antibiotics: Bacteriophages, Bacterial Cell Wall Hydrolases, and Antimicrobial Peptides. J. Appl. Microbiol. 2008, 104, 1–13.
- Gondil, V.S.; Harjai, K.; Chhibber, S. Endolysins as Emerging Alternative Therapeutic Agents to Counter Drug-Resistant Infections. Int. J. Antimicrob. Agents 2020, 55, 105844.
- Roach, D.R.; Donovan, D.M. Antimicrobial Bacteriophage-Derived Proteins and Therapeutic Applications. Bacteriophage 2015, 5, e1062590.
- Briers, Y.; Volckaert, G.; Cornelissen, A.; Lagaert, S.; Michiels, C.W.; Hertveldt, K.; Lavigne, R. Muralytic Activity and Modular Structure of the Endolysins of Pseudomonas aeruginosa Bacteriophages φKZ and EL. Mol. Microbiol. 2007, 65, 1334–1344.
- Pastagia, M.; Schuch, R.; Fischetti, V.A.; Huang, D.B. Lysins: The Arrival of Pathogen-Directed Anti-Infectives. J. Med. Microbiol. 2013, 62, 1506–1516.
- Plotka, M.; Kapusta, M.; Dorawa, S.; Kaczorowska, A.; Kaczorowski, T. Ts2631 Endolysin from the Extremophilic Thermus scotoductus Bacteriophage vB_Tsc2631 as an Antimicrobial Agent against Gram-Negative Multidrug-Resistant Bacteria. Viruses 2019, 11, 657.
- Peng, S.; You, R.; Lai, M.; Lin, N.; Chen, L.; Chang, K. Highly Potent Antimicrobial Modified Peptides Derived from the Acinetobacter baumannii Phage Endolysin LysAB2. Sci. Rep. 2017, 7, 11477.
- Yoong, P.; Schuch, R.; Nelson, D.; Fischetti, V.A. Identification of a Broadly Active Phage Lytic Enzyme with Lethal Activity against Antibiotic-Resistant Enterococcus Faecalis and Enterococcus faecium. J. Bacteriol. 2004, 186, 4808–4812.
- Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Donovan, D.M.; García, P. Enhanced Staphylolytic Activity of the Staphylococcus aureus Bacteriophage vB_SauS-phiIPLA88 HydH5 Virion-Associated Peptidoglycan Hydrolase: Fusions, Deletions, and Synergy with LysH5. Appl. Environ. Microbiol. 2012, 78, 2241–2248.
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-Encoded Virion-Associated Enzymes to Overcome the Carbohydrate Barriers during the Infection Process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119.
- Caldentey, J.; Bamford, D.H. The Lytic Enzyme of the Pseudomonas Phage Φ6. Purification and Biochemical Characterization. Biochim. Biophys. Acta (BBA)—Protein Struct. Mol. Enzymol. 1992, 1159, 44–50.
- Leiman, P.G.; Molineux, I.J. Evolution of a New Enzyme Activity from the Same Motif Fold. Mol. Microbiol. 2008, 69, 287–290.
- Cornelissen, A.; Ceyssens, P.; Krylov, V.N.; Noben, J.; Volckaert, G.; Lavigne, R. Identification of EPS-Degrading Activity within the Tail Spikes of the Novel Pseudomonas Putida Phage AF. Virology 2012, 434, 251–256.
- Pan, Y.; Lin, T.; Lin, Y.; Su, P.; Chen, C.; Hsieh, P.; Hsu, C.; Chen, C.; Hsieh, Y.; Wang, J. Identification of Capsular Types in Carbapenem-Resistant Klebsiella Pneumoniae Strains by Wzc Sequencing and Implications for Capsule Depolymerase Treatment. Antimicrob. Agents Chemother. 2015, 59, 1038–1047.
- Pan, Y.; Lin, T.; Chen, C.; Tsai, Y.; Cheng, Y.; Chen, Y.; Hsieh, P.; Lin, Y.; Wang, J. Klebsiella Phage ΦK64–1 Encodes Multiple Depolymerases for Multiple Host Capsular Types. J. Virol. 2017, 91, e02457-16.
- Carson, L.; Gorman, S.P.; Gilmore, B.F. The use of Lytic Bacteriophages in the Prevention and Eradication of Biofilms of Proteus Mirabilis and Escherichia coli. FEMS Immunol. Med. Microbiol. 2010, 59, 447–455.
- Chibeu, A.; Lingohr, E.J.; Masson, L.; Manges, A.; Harel, J.; Ackermann, H.; Kropinski, A.M.; Boerlin, P. Bacteriophages with the Ability to Degrade Uropathogenic Escherichia Coli Biofilms. Viruses 2012, 4, 471–487.
- Meng, X.; Shi, Y.; Ji, W.; Meng, X.; Zhang, J.; Wang, H.; Lu, C.; Sun, J.; Yan, Y. Application of a Bacteriophage Lysin to Disrupt Biofilms Formed by the Animal Pathogen Streptococcus Suis. Appl. Environ. Microbiol. 2011, 77, 8272–8279.
- Topka-Bielecka, G.; Dydecka, A.; Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Węgrzyn, G.; Węgrzyn, A. Bacteriophage-Derived Depolymerases against Bacterial Biofilm. Antibiotics 2021, 10, 175.
- Gutiérrez, D.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; García, P. Bacteriophages as Weapons against Bacterial Biofilms in the Food Industry. Front. Microbiol. 2016, 7, 825.
- Saier, M.H., Jr.; Reddy, B.L. Holins in Bacteria, Eukaryotes, and Archaea: Multifunctional Xenologues with Potential Biotechnological and Biomedical Applications. J. Bacteriol. 2015, 197, 7–17.
- Wang, I.; Smith, D.L.; Young, R. Holins: The Protein Clocks of Bacteriophage Infections. Annu. Rev. Microbiol. 2000, 54, 799–825.
- Song, J.; Niu, W.; Wu, R.; Wang, J.; Lei, L.; Han, W.; Gu, J. The Phage Holin HolGH15 Exhibits Potential as an Antibacterial Agent to Control Listeria Monocytogenes. Foodborne Pathog. Dis. 2021, 18, 574–581.
- Vilcinskas, A. Evolutionary Plasticity of Insect Immunity. J. Insect Physiol. 2013, 59, 123–129.
- Wu, Q.; Patočka, J.; Kuča, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461.
- Dutta, P.; Sahu, R.K.; Dey, T.; Lahkar, M.D.; Manna, P.; Kalita, J. Beneficial Role of Insect-Derived Bioactive Components against Inflammation and its Associated Complications (Colitis and Arthritis) and Cancer. Chem. Biol. Interact. 2019, 313, 108824.
- Buonocore, F.; Fausto, A.M.; Pelle, G.D.; Roncevic, T.; Gerdol, M.; Picchietti, S. Attacins: A Promising Class of Insect Antimicrobial Peptides. Antibiotics 2021, 10, 212.
- Zahedifard, F.; Lee, H.; No, J.H.; Salimi, M.; Seyed, N.; Asoodeh, A.; Rafati, S. Comparative Study of Different Forms of Jellein Antimicrobial Peptide on Leishmania Parasite. Exp. Parasitol. 2020, 209, 107823.
More
