Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Jessie Wu and Version 1 by Zhiyin Song.
Mitochondrion harbors its own DNA (mtDNA), which encodes many critical proteins for the assembly and activity of mitochondrial respiratory complexes. mtDNA is packed by many proteins to form a nucleoid that uniformly distributes within the mitochondrial matrix, which is essential for mitochondrial functions. Defects or mutations of mtDNA result in a range of diseases. Damaged mtDNA could be eliminated by mitophagy, and all paternal mtDNA are degraded by endonuclease G or mitophagy during fertilization.
- mtDNA
- mitophagy
1. mtDNA Structure
1. mtDNA Structure
The structure of mtDNA is significantly different from that of nDNA; however, similar to the bacterial chromosome, mtDNA forms a closed circle doubled-stranded DNA in nearly all metazoa [19][1]. The sense strand and antisense strand of mtDNA are named a heavy (H) strand and a light (L) strand. In human cells, mtDNA consists of 16,569 base pairs, and encodes 37 genes, including 13 polypeptides, two ribosomal RNAs, and 22 tRNAs [6,20][2][3]. One polypeptide (ND6) and eight tRNAs are located on the L strand; the other 12 polypeptides, two rRNAs, and 14 tRNAs are encoded by the H strand. mtDNA also contains a noncoding region, which is called a displacement loop (D-loop), and harbors almost all the known mtDNA replication and transcription [21][4]. The 13 polypeptides are the core subunit of the oxidative phosphorylation (OXPHOS) complexes I, III, IV, and V, and are essential for OXPHOS activity. Mitochondrial rRNAs and tRNAs constitute a machine for the synthesis of 13 peptides.
2. mtDNA Mutation and Human Diseases
2. mtDNA Mutation and Human Diseases
mtDNA is susceptible to be attacked by oxygen free radicals, and tends to develop somatic mutations due to the lack of protection by histones [22,23][5][6]. mtDNA is located in the mitochondrial matrix, and is in close proximity to the respiratory chains [20[3][6],23], which are the main source of the reactive oxygen species (ROS). mtDNA encodes the core subunit of OXPHOS that produces the vast majority of cellular ATP. Excessive mtDNA mutations could result in the dysfunction of OXPHOS, which subsequently leads to diseases associated with mitochondrial function. In fact, many diseases have been found to be associated with mtDNA mutations, and most maternal mtDNA diseases can transmit to their offspring due to the feature of matrilineal inheritance in mtDNA [24][7].
Since the first human mtDNA mutation was described in 1988 [25][8], several mtDNA mutations and the associated mtDNA diseases have been identified. The obvious feature of mtDNA diseases is characterized by the presence of various neurological features [19][1]. Kearns–Sayre syndrome (KSS) and Leber’s hereditary optic neuropathy (LHON) are the early identified syndromes associated with mtDNA mutation [26,27][9][10]. KSS is associated with progressive myopathy, ophthalmoplegia, and cardiomyopathy, which is caused by single, large-scale deletions [25,26][8][9]. LHON is an optic neuropathy that is caused by mtDNA point mutations (m.3460G > A, m.11778G > A, and m.14484T > C) [27,28,29][10][11][12]. The point mutation of ATP6 (m.8993T > C or 8993T > G), which is the core subunit of OXPHOS protein complex V, contributes to Leigh syndrome (LS), which is also known as subacute necrotizing encephalomyelopathy [30,31][13][14]. Myoclonic epilepsy with ragged-red fibers (MERRF), which is a severe neuromuscular disorder accompanied by symptoms of myoclonic epilepsy, myopathy, dementia, or ataxia, is caused by the point mutation of tRNA [32,33][15][16].
Additional, mtDNA mutations are associated with other human diseases, including diabetes, Alzheimer’s disease (AD), Parkinson’s disease (PD), and cancer. Diabetes is one of the most common chronic disorders. mtDNA point mutations (m.3242A > G) and the 10.4-kb deletion of mtDNA are associated with diabetes and deafness, and the mutations are maternally inherited [34,35][17][18]. It is hypothesized that mtDNA mutations accumulate over time, which plays a central role in the process of aging and related neurodegeneration [19][1]. In fact, there is already a lot of evidence that demonstrates that mtDNA mutations are indeed associated with aging, Parkinson’s disease, and Alzheimer’s disease. Recent evidence suggests that dysregulated mitochondrial dynamics and mutations caused by mtDNA replication can lead to aging, and the increasing mtDNA mutation rates increase the aging rate and provide an aging clock [36][19]. A high level of deleted mtDNA has been found in the substantia nigra neurons of patients with aging and Parkinson’s disease [37][20]. Parkinson’s disease is a neurodegenerative disease that is characterized by the loss of dopamine neurons in the substantia nigra of the brain and the accumulation of α-synuclein [38][21]. Alzheimer’s disease, another neurodegenerative disease, is associated with heteroplasmic mtDNA mutations [39][22]. In addition, tumors and mtDNA mutations are also inextricably linked. mtDNA mutations contribute to tumorigenicity. ND3 gene mutation (m.G10398A) had been found to increase the risk of invasive breast cancer in African-American women [40][23]. Further data demonstrate that both germ-line and somatic mtDNA mutations contribute to prostate cancer, and about 11% of all prostate cancer patients harbored mt-CO1 (mitochondrially encoded cytochrome c oxidase I) mutations [41][24]. Additionally, the pathogenic mtDNA ATP6 T8993G germ-line mutation was found to generate tumors that were seven times larger than the wild type (T8993T) [41][24].
3. mtDNA Distribution
3. mtDNA Distribution
The mitochondrion is a highly dynamic double-membrane organelle that forms a well-distributed network in the majority of mammalian cell types. mtDNA is located in the mitochondrial matrix, associated with the mitochondrial inner membrane, and distributed throughout the mitochondrial network [20][3]. Each mitochondrion contains one or more mtDNA molecules [6][2]. In proliferative cells, mtDNA is replicated, separated, and distributed equally to daughter cells, which are dependent on mitochondrial dynamics. In addition, the mitochondrial membrane structure and membrane composition are also involved in mtDNA attachment and distribution [20][3].
3.1. mtDNA Distribution and Mitochondrial Dynamics
3.1. mtDNA Distribution and Mitochondrial Dynamics
Mitochondria continuously undergo fusion and fission, which are essential for cell metabolic activities, as well as mtDNA distribution in mitochondria. Mitochondrial fusion and fission, the two opposite processes, are both mediated by large GTPases proteins, which are conserved in yeast, flies, and mammals [42][25]. Mitochondrial fusion is mediated by three GTPases proteins: Mitofusin 1 (Mfn1), Mitofusin 2 (Mfn2), and Optic Atrophy 1 (OPA1) [43,44][26][27]. As the feature of a double membrane, mitochondrial fusion is a two-step process requiring outer-membrane fusion followed by inner-membrane fusion [1][28]. Mfn1 and Mfn2 regulate the mitochondrial outer membrane fusion, and OPA1 is involved in mitochondrial inner membrane fusion [45][29]. A deficiency of fusion results in severe mitochondrial fragmentation and is associated with a range of human diseases [46,47][30][31]. The mutation of Mfn2 causes Charcot–Marie–Tooth disease type 2A in human, which is a common inherited peripheral neuropathy [47,48][31][32]. The dysfunction of OPA1 is associated with dominant optic atrophy (DOA), which is an optic neuropathy caused by the degeneration of retinal ganglion cells [1,49,50][28][33][34]. Mitochondrial fission is regulated by Drp1, a cytosolic dynamic protein, which is recruited to mitochondria from the cytosol, forms spirals around the mitochondria, and then constricts it by hydrolyzing GTP to mediate mitochondrial scission [1,51][28][35].
Mitochondria and mtDNA are highly dynamic [52][36]. mtDNA are distributed throughout the mitochondrial network [53][37], which is important for the uniform distribution of mtDNA-encoded proteins in mitochondria. Mitochondrial dynamics greatly influence the distribution and maintenance of mtDNA [54][38]. A deficiency in mitochondrial fusion has a profound effect on mtDNA (Figure 1A). It has been demonstrated Mfn1 and Mfn2 conditional knock-out mice in muscle result in muscle atrophy, mitochondrial dysfunction, and severe mtDNA depletion [55][39]. OPA1 mediates the fusion of the mitochondrial inner membrane, and regulates cristae remodeling and cytochrome c release during apoptosis [56,57,58][40][41][42]. In addition, OPA1 mutations in patients lead to multiple deletions of mtDNA in their skeletal muscle [59][43], and one isoform of OPA1 was associated with mtDNA replication, distribution, and maintenance [60][44]. Mitochondrial fission also plays an essential role in mtDNA distribution. The deficiency of mitochondrial fission caused by the loss of Drp1 leads to hyperfused mitochondria and enlarged mtDNA nucleoids characterized by mtDNA accumulation [54,61,62][38][45][46]. Mitochondrial fusion promotes complementation between two mitochondria, including mtDNA [42,63][25][47]; mitochondrial fission separates mtDNAs into two divided mitochondria, and also contributes to a chance for a mitochondrion to re-fuse with another part of the mitochondrial network (Figure 1A). Therefore, mtDNA are distributed throughout the network by continuous fusion and fission [54][38].
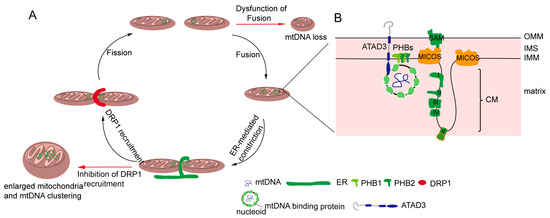
Figure 1. Regulation of the distribution of mitochondria DNA (mtDNA). (A) Mitochondrial dynamics regulate mtDNA. Mitochondrial fission and mtDNA segregation happened synchronously, and occur at the ER and mitochondrial contact site. Upon fission, the endoplasmic reticulum (ER) wraps the mitochondria, and then the cytosolic dynamic protein Drp1 is recruited to mediated mitochondria division. Blocking the fission leads to enlarged mitochondria and an mtDNA cluster. Mitochondrial fusion allows for two mitochondrial exchange substances, including mtDNA. The dysfunction of fusion leads to mtDNA deletion. (B) The mitochondrial inner membrane is involved in mtDNA distribution. Certain mitochondrial inner membrane proteins such as prohibitins and ATPase family AAA domain-containing protein 3 (ATAD3) are mtDNA-binding proteins. In addition, mtDNA nucleoid contacts with the mitochondrial cristae junction, and MICOS complex and Sam50, which are involved in the maintenance of the cristae structure, regulate mtDNA distribution.
The distribution of mtDNA is tightly interlinked with the dynamics of mitochondria, but the mechanisms of mtDNA distribution throughout the mitochondrial network are poorly understood. Recent evidence shows the close proximity between mtDNA and the sites of Drp1-dependent mitochondrial fission, which is highly conserved in yeast and mammalian cells [61,64,65][45][48][49]. In yeast and mammalian cells, mitochondrial division occurs at the endoplasmic reticulum (ER) and mitochondria contact sites (Figure 1A), in which the ER wraps around the mitochondria; then, Drp1 is recruited and assembled around mitochondria [66,67][50][51]. Moreover, the majority of ER-linked mitochondrial division events occur adjacent to nucleoids [20,65][3][49]. Following mtDNA replication, ER-linked mitochondrial fission occurs between the replicated mtDNAs, which locate at newly generated mitochondrial tips after scission [53,64,65][37][48][49]. Localizing mtDNA to the newly formed mitochondrial tips could transport mtDNA to the distal parts of cell, and further fuse with other mitochondria to drive mtDNA distribution. The mechanism can explain how mtDNA is equivalently distributed in cells and how mtDNA is distributed into mitochondria following mtDNA replication.
3.2. mtDNA Distribution and Inner Membrane Structure
3.2. mtDNA Distribution and Inner Membrane Structure
The structure of the inner mitochondrial membrane (IMM) is divided into two morphologically and presumably functionally distinct subdomains: the inner boundary membrane (IBM), which is closely opposed to the outer mitochondrial membrane (OMM), and the cristae membrane (CM), which protrudes into the matrix [20,68][3][52]. The IBM comes into close contact with the OM by the protein transport complexes [68,69,70][52][53][54]. The CM is formed by the invaginations of the IBM, and is enriched in respiratory chain complexes and some small molecules and metabolites [68,71][52][55]. There is another substructure of the inner membrane—the cristae junction—that connects the IBM with the CM [72,73][56][57]. It has been reported mtDNA is associated with the IMM, and mtDNA is frequently observed intertwined into cristae [20][3]. Therefore, there may be several IMM factors regulating mtDNA distribution. Indeed, it has been found that the MICOS (mitochondrial contact site and cristae junction organizing system) locates at the cristae junction and is involved in regulating the inner mitochondrial membrane cristae junction [71,74,75][55][58][59]. In yeast, MIC60 (Fcj1) and Mic10 (Mos10), two key components of the MICOS, regulate mtDNA nucleoid size and distribution [76][60]. Deficiencies in the two proteins result in the formation of large mtDNA nucleoids and giant spherical mitochondria [76][60]. Consistently, researchers have found that MIC60 (IMMT) knockdown led to alterations of mitochondrial tubular morphology to giant spherical mitochondria and the disorganization and clustering of nucleoids in mammalian cells (Figure 1A) [77][61]. Sam50, a MICOS-interacting protein in mammalian cells, is located at the outer mitochondrial membrane [78][62]. The loss of Sam50 results in the disorganization of cristae and large spherical mitochondria, and also leads to enlarged mtDNA nucleoids, which protect mtDNA from clearance by mitophagy [79][63]. However, how the mitochondrial inner membrane regulates mtDNA organization and distribution remains unknown. It has been hypothesized that cristae junctions contribute to maintaining proper internal membrane compartmentalization, and the loss of these junctions leads to clustering and the missegregation of mtDNA nucleoids due to the loss of proper compartmental localization of the mtDNA within the mitochondrial tubules [71][55].
3.3. mtDNA Distribution and Cholesterol
3.3. mtDNA Distribution and Cholesterol
Cholesterol is a composition of lipid rafts, and contributes to being a dynamic glue that keeps the raft assembly together [80,81][64][65]. Recent data demonstrate that the human mtDNA–protein complex colocalizes with the cholesterol-rich membrane [82][66]. Additional, cholesterol is also rich at the site of the ER-associated mitochondrial membrane (MAM), which is involved in mtDNA distribution and segregation [20,83,84][3][67][68]. Thus, it is possible that cholesterol is associated with the distribution of mitochondrial nucleoids. ATAD3 (ATPase family AAA domain-containing protein 3), locating at the mitochondrial inner membrane, is colocalized with mitochondrial nucleoids in mammalian cells by binding to the D-loop of mtDNA (Figure 1B) [85,86][69][70]. A deficiency of ATAD3 in cells results in the disorganization of mitochondrial nucleoids, which is also found in the mouse model and in patients with pathogenic mutations in ATAD3 [87,88][71][72]. Furthermore, ATAD3 is involved in regulating cholesterol metabolism [87,88][71][72]. Therefore, it seems that ATAD3 regulates mtDNA maintenance by regulating cholesterol metabolism.
References
- Li, Z.; Zhou, T.; Chuang, C.-C. The Consequences of Damaged Mitochondrial DNA. In Mitochondrial Mechanisms of Degeneration and Repair in Parkinson’s Disease; Buhlman, L.M., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 49–61.
- Mishra, P.; Chan, D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 634–646.
- Nicholls, T.J.; Gustafsson, C.M. Separating and Segregating the Human Mitochondrial Genome. Trends Biochem. Sci. 2018, 43, 869–881.
- Sbisa, E.; Tanzariello, F.; Reyes, A.; Pesole, G.; Saccone, C. Mammalian mitochondrial D-loop region structural analysis: Identification of new conserved sequences and their functional and evolutionary implications. Gene 1997, 205, 125–140.
- Richter, C. Oxidative damage to mitochondrial DNA and its relationship to ageing. Int. J. Biochem. Cell Biol. 1995, 27, 647–653.
- Shokolenko, I.; Venediktov, N.; Bochkareva, A.; Wilson, G.L.; Alexeyev, M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009, 37, 2539–2548.
- van Oven, M.; Kayser, M. Updated Comprehensive Phylogenetic Tree of Global Human Mitochondrial DNA Variation. Hum. Mutat. 2009, 30, E386–E394.
- Holt, I.J.; Harding, A.E.; Morganhughes, J.A. Deletions of Muscle Mitochondrial-DNA in Patients with Mitochondrial Myopathies. Nature 1988, 331, 717–719.
- Zeviani, M.; Moraes, C.T.; DiMauro, S.; Nakase, H.; Bonilla, E.; Schon, E.A.; Rowland, L.P. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology 1988, 38, 1339.
- Wallace, D.; Singh, G.; Lott, M.T.; Hodge, J.A.; Schurr, T.G.; Lezza, A.M.; Elsas, L.J.; Nikoskelainen, E.K. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science 1988, 242, 1427–1430.
- Howell, N.; Bindoff, L.A.; McCullough, D.A.; Kubacka, I.; Poulton, J.; Mackey, D.; Taylor, L.; Turnbull, D.M. Leber hereditary optic neuropathy: Identification of the same mitochondrial ND1 mutation in six pedigrees. Am. J. Hum. Genet. 1991, 49, 939–950.
- Johns, D.R.; Neufeld, M.J.; Park, R.D. An ND-6 mitochondrial DNA mutation associated with Leber hereditary optic neuropathy. Biochem. Biophys. Res. Commun. 1992, 187, 1551–1557.
- de Vries, D.D.; van Engelen, B.G.; Gabreëls, F.J.; Ruitenbeek, W.; van Oost, B.A. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh’s syndrome. Ann. Neurol. 1993, 34, 410–412.
- Holt, I.J.; Harding, A.E.; Petty, R.K.; Morgan-Hughes, J.A. A New Mitochondrial Disease Associated with Mitochondrial-DNA Heteroplasmy. Am. J. Hum. Genet. 1990, 46, 428–433.
- Shoffner, J.M.; Lott, M.T.; Lezza, A.M.; Seibel, P.; Ballinger, S.W.; Wallace, D.C. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 1990, 61, 931–937.
- Silvestri, G.; Moraes, G.T.; Shanske, S.; Oh, S.J.; Di Mauro, S. A new mtDNA mutation in the tRNA(Lys) gene associated with myoclonic epilepsy and ragged-red fibers (MERRF). Am. J. Hum. Genet. 1992, 51, 1213–1217.
- Ballinger, S.W.; Shoffner, J.M.; Hedaya, E.V.; Trounce, I.; Polak, M.A.; Koontz, D.A.; Wallace, D.C. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat. Genet. 1992, 1, 11–15.
- Kadowaki, T.; Kadowaki, H.; Mori, Y.; Tobe, K.; Sakuta, R.; Suzuki, Y.; Tanabe, Y.; Sakura, H.; Awata, T.; Goto, Y.; et al. A Subtype of Diabetes Mellitus Associated with a Mutation of Mitochondrial DNA. N. Engl. J. Med. 1994, 330, 962–968.
- Wallace, D.C. Mitochondrial DNA Mutations in Disease and Aging. Environ. Mol. Mutagen. 2010, 51, 440–450.
- Bender, A.; Krishnan, K.J.; Morris, C.M.; Taylor, G.A.; Reeve, A.K.; Perry, R.H.; Jaros, E.; Hersheson, J.S.; Betts, J.; Klopstock, T.; et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006, 38, 515.
- Dauer, W.; Przedborski, S. Parkinson’s Disease: Mechanisms and Models. Neuron 2003, 39, 889–909.
- Howell, N.; Elson, J.L.; Chinnery, P.F.; Turnbull, D.M. mtDNA mutations and common neurodegenerative disorders. Trends Genet. 2005, 21, 583–586.
- Brandon, M.; Baldi, P.; Wallace, D.C. Mitochondrial mutations in cancer. Oncogene 2006, 25, 4647–4662.
- Petros, J.A.; Baumann, A.K.; Ruiz-Pesini, E.; Amin, M.B.; Sun, C.Q.; Hall, J.; Lim, S.; Issa, M.M.; Flanders, W.D.; Hosseini, S.H.; et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 719–724.
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065.
- Chan, D.C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006, 22, 79–99.
- Chen, H.; Chan, D.C. Mitochondrial dynamics—Fusion, fission, movement, and mitophagy—In neurodegenerative diseases. Hum. Mol. Genet. 2009, 18, R169–R176.
- Mishra, P.; Chan, D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016, 212, 379–387.
- Song, Z.; Ghochani, M.; McCaffery, J.M.; Frey, T.G.; Chan, D.C. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell 2009, 20, 3525–3532.
- Chen, H.; Ghochani, M.; McCaffery, J.M.; Frey, T.G.; Chan, D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003, 160, 189–200.
- Detmer, S.A.; Chan, D.C. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J. Cell Biol. 2007, 176, 405–414.
- Zuchner, S.; Mersiyanova, I.V.; Muglia, M.; Bissar-Tadmouri, N.; Rochelle, J.; Dadali, E.L.; Zappia, M.; Nelis, E.; Patitucci, A.; Senderek, J.; Parman, Y.; et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 2004, 36, 449–451.
- Delettre, C.; Lenaers, G.; Griffoin, J.M.; Gigarel, N.; Lorenzo, C.; Belenguer, P.; Pelloquin, L.; Grosgeorge, J.; Turc-Carel, C.; Perret, E.; et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000, 26, 207–210.
- Alexander, C.; Votruba, M.; Pesch, U.E.; Thiselton, D.L.; Mayer, S.; Moore, A.; Rodriguez, M.; Kellner, U.; Leo-Kottler, B.; Auburger, G.; et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000, 26, 211–215.
- Smirnova, E.; Griparic, L.; Shurland, D.L.; van der Bliek, A.M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 2001, 12, 2245–2256.
- Malka, F.; Lombes, A.; Rojo, M. Organization, dynamics and transmission of mitochondrial DNA: Focus on vertebrate nucleoids. Biochim. Biophys. Acta 2006, 1763, 463–472.
- Osman, C.; Noriega, T.R.; Okreglak, V.; Fung, J.C.; Walter, P. Integrity of the yeast mitochondrial genome, but not its distribution and inheritance, relies on mitochondrial fission and fusion. Proc. Natl. Acad. Sci. USA 2015, 112, E947–E956.
- Jayashankar, V.; Rafelski, S.M. Integrating mitochondrial organization and dynamics with cellular architecture. Curr. Opin. Cell Biol. 2014, 26, 34–40.
- Chen, H.C.; Vermulst, M.; Wang, Y.E.; Chomyn, A.; Prolla, T.A.; McCaffery, J.M.; Chan, D.C. Mitochondrial Fusion Is Required for mtDNA Stability in Skeletal Muscle and Tolerance of mtDNA Mutations. Cell 2010, 141, 280–289.
- Cipolat, S.; Rudka, T.; Hartmann, D.; Costa, V.; Serneels, L.; Craessaerts, K.; Metzger, K.; Frezza, C.; Annaert, W.; D’Adamio, L.; et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 2006, 126, 163–175.
- Olichon, A.; Baricault, L.; Gas, N.; Guillou, E.; Valette, A.; Belenguer, P.; Lenaers, G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003, 278, 7743–7746.
- Frezza, C.; Cipolat, S.; Martins de Brito, O.; Micaroni, M.; Beznoussenko, G.V.; Rudka, T.; Bartoli, D.; Polishuck, R.S.; Danial, N.N.; de Strooper, B.; et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 2006, 126, 177–189.
- Amati-Bonneau, P.; Valentino, M.L.; Reynier, P.; Gallardo, M.E.; Bornstein, B.; Boissière, A.; Campos, Y.; Rivera, H.; de la Aleja, J.G.; Carroccia, R.; et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy plus phenotypes. Brain 2008, 131, 338–351.
- Elachouri, G.; Vidoni, S.; Zanna, C.; Pattyn, A.; Boukhaddaoui, H.; Gaget, K.; Yu-Wai-Man, P.; Gasparre, G.; Sarzi, E.; Delettre, C.; et al. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011, 21, 12–20.
- Ban-Ishihara, R.; Ishihara, T.; Sasaki, N.; Mihara, K.; Ishihara, N. Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c. Proc. Natl. Acad. Sci. USA 2013, 110, 11863–11868.
- Parone, P.A.; Da Cruz, S.; Tondera, D.; Mattenberger, Y.; James, D.I.; Maechler, P.; Barja, F.; Martinou, J.C. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE 2008, 3, e3257.
- Nakada, K.; Inoue, K.; Ono, T.; Isobe, K.; Ogura, A.; Goto, Y.I.; Nonaka, I.; Hayashi, J.I. Inter-mitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat. Med. 2001, 7, 934–940.
- Murley, A.; Lackner, L.L.; Osman, C.; West, M.; Voeltz, G.K.; Walter, P.; Nunnari, J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife 2013, 2, e00422.
- Lewis, S.C.; Uchiyama, L.F.; Nunnari, J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 2016, 353, aaf5549.
- Friedman, J.R.; Lackner, L.L.; West, M.; DiBenedetto, J.R.; Nunnari, J.; Voeltz, G.K. ER tubules mark sites of mitochondrial division. Science 2011, 334, 358–362.
- Korobova, F.; Ramabhadran, V.; Higgs, H.N. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 2013, 339, 464–467.
- Vogel, F.; Bornhövd, C.; Neupert, W.; Reichert, A.S. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 2006, 175, 237–247.
- Yamamoto, H.; Esaki, M.; Kanamori, T.; Tamura, Y.; Nishikawa, S.; Endo, T. Tim50, a new subunit of the TIM23 complex that links mitochondrial protein translocation across the outer membrane and that across the inner membrane. Mol. Biol. Cell 2002, 13, 128a.
- Donzeau, M.; Káldi, K.; Adam, A.; Paschen, S.; Wanner, G.; Guiard, B.; Bauer, M.F.; Neupert, W.; Brunner, M. Tim23 links the inner and outer mitochondrial membranes. Cell 2000, 101, 401–412.
- Schorr, S.; van der Laan, M. Integrative functions of the mitochondrial contact site and cristae organizing system. Semin. Cell Dev. Biol. 2018, 76, 191–200.
- Cogliati, S.; Enriquez, J.A.; Scorrano, L. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem. Sci. 2016, 41, 261–273.
- Renken, C.; Siragusa, G.; Perkins, G.; Washington, L.; Nulton, J.; Salamon, P.; Frey, T.G. A thermodynamic model describing the nature of the crista junction: A structural motif in the mitochondrion. J. Struct. Biol. 2002, 138, 137–144.
- van der Laan, M.; Horvath, S.E.; Pfanner, N. Mitochondrial contact site and cristae organizing system. Curr. Opin. Cell Biol. 2016, 41, 33–42.
- Pfanner, N.; van der Laan, M.; Amati, P.; Capaldi, R.A.; Caudy, A.A.; Chacinska, A.; Darshi, M.; Deckers, M.; Hoppins, S.; Icho, T.; et al. Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J. Cell Biol. 2014, 204, 1083–1086.
- Itoh, K.; Tamura, Y.; Iijima, M.; Sesaki, H. Effects of Fcj1-Mos1 and mitochondrial division on aggregation of mitochondrial DNA nucleoids and organelle morphology. Mol. Biol. Cell 2013, 24, 1842–1851.
- Li, H.; Ruan, Y.; Zhang, K.; Jian, F.; Hu, C.; Miao, L.; Gong, L.; Sun, L.; Zhang, X.; Chen, S.; et al. Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ. 2016, 23, 380–392.
- Kozjak-Pavlovic, V. The MICOS complex of human mitochondria. Cell Tissue Res. 2017, 367, 83–93.
- Jian, F.; Chen, D.; Chen, L.; Yan, C.; Lu, B.; Zhu, Y.; Chen, S.; Shi, A.; Chan, D.C.; Song, Z. Sam50 Regulates PINK1-Parkin-Mediated Mitophagy by Controlling PINK1 Stability and Mitochondrial Morphology. Cell Rep. 2018, 23, 2989–3005.
- Simons, K.; Ehehalt, R. Cholesterol, lipid rafts, and disease. J. Clin. Invest. 2002, 110, 597–603.
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39.
- Gerhold, J.M.; Cansiz-Arda, Ş.; Lõhmus, M.; Engberg, O.; Reyes, A.; van Rennes, H.; Sanz, A.; Holt, I.J.; Cooper, H.M.; Spelbrink, J.N. Human Mitochondrial DNA-Protein Complexes Attach to a Cholesterol-Rich Membrane Structure. Sci. Rep. 2015, 5, 15292.
- Area-Gomez, E.; del Carmen Lara Castillo, M.; Tambini, M.D.; Guardia-Laguarta, C.; de Groof, A.J.C.; Madra, M.; Ikenouchi, J.; Umeda, M.; Bird, T.D.; Sturley, S.L.; et al. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 2012, 31, 4106–4123.
- Fujimoto, M.; Hayashi, T.; Su, T.P. The role of cholesterol in the association of endoplasmic reticulum membranes with mitochondria. Biochem. Biophys. Res. Commun. 2012, 417, 635–639.
- He, J.; Mao, C.C.; Reyes, A.; Sembongi, H.; Di Re, M.; Granycome, C.; Clippingdale, A.B.; Fearnley, I.M.; Harbour, M.; Robinson, A.J.; et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007, 176, 141–146.
- Hubstenberger, A.; Merle, N.; Charton, R.; Brandolin, G.; Rousseau, D. Topological analysis of ATAD3A insertion in purified human mitochondria. J. Bioenerg. Biomembr. 2010, 42, 143–150.
- Desai, R.; Frazier, A.E.; Durigon, R.; Patel, H.; Jones, A.W.; Dalla Rosa, I.; Lake, N.J.; Compton, A.G.; Mountford, H.S.; Tucker, E.J.; et al. ATAD3 gene cluster deletions cause cerebellar dysfunction associated with altered mitochondrial DNA and cholesterol metabolism. Brain 2017, 140, 1595–1610.
- Peralta, S.; Goffart, S.; Williams, S.L.; Diaz, F.; Garcia, S.; Nissanka, N.; Area-Gomez, E.; Pohjoismäki, J.; Moraes, C.T. ATAD3 controls mitochondrial cristae structure in mouse muscle, influencing mtDNA replication and cholesterol levels. J. Cell Sci. 2018, 131, jcs217075.
More
