Biocontrol of a pioneer fungus of esca, a frequent grapevine trunk disease, was investigated by deciphering the tripartite interaction between this trunk-esca pathogen (
Phaeomoniella chlamydospora
), grapevine and the biocontrol-oomycete,
Pythium oligandrum
. Grapevine-defense was enhanced in response to
P. chlamydospora
attacks, with
P. oligandrum
acting as a plant-systemic resistance inducer, promoting jasmonic/ethylene signaling pathways and grapevine priming.
P. chlamydospora
pathogenicity genes, such as those related to secondary metabolite biosynthesis, carbohydrate-active enzymes and transcription regulators, were also affected in their expression. Shifts in grapevine responses and key-fungal functions were associated with the reduction of
P. chlamydospora
wood necroses. This study provides evidence of grapevine-wood fungal pathogen transcriptional changes induced by a root biocontrol agent,
P. oligandrum
, in which there is no contact between the two microorganisms.
- transcriptomic analyses
- Biocontrol
- grapevine trunk disease
- Pythium oligandrum
1. Introduction
Applying biocontrol agents (BCAs) to plants can be useful to control diseases, particularly when chemical pesticides have been banned or do not exist [1,2][1][2]. This is the case for grapevine trunk diseases (GTDs), which have increased worldwide over the last two decades following the ban, in Europe in the early 2000s, of sodium arsenite, due to its toxic effect on the environment and humans [1,3][1][3]. GTDs, especially esca, cause considerable economic losses to the viticulture sector [4], with decreases in plant productivity and longevity, and wine quality alteration [5,6][5][6]. The worldwide annual incidence and cost for the replacement of dead plants due to GTDs is very high. In Italy, for instance, depending on the cultivars, the incidence of GTDs ranged from 8% to 19% and around 10% in Spain; in France, approximately 12% of French vineyards are unproductive during the period of 2012-2017 [7], with losses, in 2014, estimated at around € 1 billion.
In order to control esca, various beneficial microorganisms, used as BCAs, have been tested to control the fungal pathogens associated with this disease [1,3,8][1][3][8]. Two biocontrol products called Esquive WP® and Vintec® based on two different strains of Trichoderma atroviride, I-1237 and SC1, respectively, have been registered to control esca on the adult grapevine [9,10][9][10]. However, their protection levels usually vary from year to year. Consequently, the few BCAs available against GTDs, particularly esca, need their levels of protection to be optimized. Improving our understanding of the molecular mechanisms involved in a defined grapevine, esca-pathogen and biocontrol agent interaction would facilitate the development of this control method.
Among the potential microbial BCAs, we have selected here the oomycete, Pythium oligandrum, because it is naturally present in the rhizosphere of many plants, including vineyards [11], and has shown potential in controlling many plant pathogens [12,13][12][13]. As regards the plant protection provided by P. oligandrum, it results from: (1) the direct effect of the oomycete on the pathogens by destroying them via mycoparasitism, antibiosis and/or competition for nutrient elements and (2) the indirect effect P. oligandrum has on plants by inducing host resistance. Scientists generally considered plant-induced resistance as the main mode of action of P. oligandrum [13]. Three elicitin-like proteins produced by P. oligandrum, such as oligandrin, POD1 and POD2, are also able to trigger this host-resistance when they are applied on plants.
Regarding grapevines, previous studies showed that treating Vitis vinifera L. roots with P. oligandrum was associated with a reduction in Botrytis cinerea infection at the leaf level [14]. It also reduced the wood attacks of two fungal pathogens involved in GTDs [15], Phaeomoniella chlamydospora and Neofusicoccum parvum [16,17][16][17]. Hence, the use of a BCA to control esca could well prove an interesting approach for growers in developing sustainable viticulture.
Various molecular aspects of a grapevine, esca-pathogen and biocontrol agent interaction will be investigated here, since little is known about the molecular events associated with GTD pathogens. The complete genome sequencing of several GTD pathogens [18,19,20][18][19][20] has also paved the way for obtaining molecular insights about the genes involved in the pathological process. Moreover, this enables improved understanding of the plant resistance induced by the BCA.
2. Evaluation of Grapevine Transcriptomic Responses
Differentially expressed (DE) genes in the plant wood tissues were investigated using a time- frame approach with V. vinifera microarrays (0- vs. 14-dpi corresponding to 2 h and 14 days after pathogen infection).
2.1. Grapevine Trunk Transcriptional Response to P. oligandrum Root Colonization
Transcriptomic analysis of P. oligandrum-treated plants at 0- vs. 14-dpi revealed that a total of 189 genes were differentially expressed. Seventy percent of these genes (green genes) were overexpressed at 0-dpi. A few functional categories showed statistical significance (Figure 1). Stress-associated genes showed overexpression at 0-dpi (PR-proteins and heat shock protein). On the contrary, polyamine metabolism (two S-adenosylmethionine decarboxylase genes, SAM) and three GCN5-related-N-acetyltransferase associated genes showed overexpression at 14-dpi. As regards the hormone metabolism category, whereas DE genes related to ethylene (ET), gibberellin and salicylic acid were induced at 0-dpi, those concerning cytokinin were overexpressed at 14-dpi.
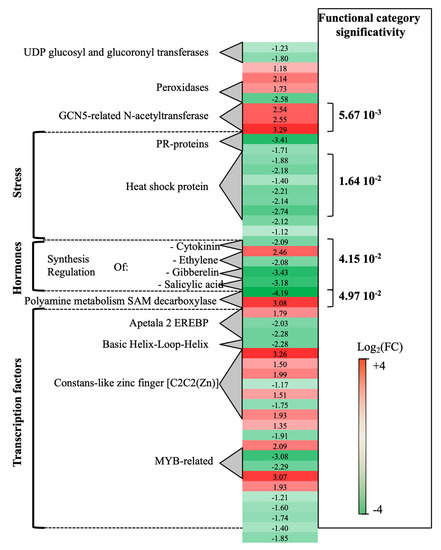
Figure 1. Global aspects of the grapevine transcriptome evolution from 0- to 14-dpi according to the treatments. Major functional categories obtained after Limma analysis and Mapman classification for root-inoculated grapevine with Pythium oligandrum, in the absence of pathogen infection. The red color means overexpression at 14-dpi and the green color means overexpression at 0-dpi.
Other important functional categories, albeit of lesser statistical significance, concerned transcriptional regulation factors: apetala2 ethylene-responsive element binding protein transcription factor family (AP2-EREBP), which was overexpressed, at 0-dpi (3 genes), and basic helix-loop-helix type transcription factor (bHLH), overexpressed at 14-dpi (1 gene).
2.2. Transcriptomic Changes in Grapevine Wood Tissues Following P. chlamydospora or P. oligandrum + P. chlamydospora inoculations
In order to evaluate grapevine transcriptomic changes, plant responses to the different treatments (mock inoculation, P. chlamydospora and P. oligandrum + P. chlamydospora) were compared. DE genes between 0- and 14-dpi were obtained using Limma analysis (p < 0.01), which gave the following results: mock treatment (1371 genes), pathogen treatment (2235 genes) and pathogen treatment in the presence of the root BCA (1858 genes). The number of specific and common DE genes detected is shown in the Venn diagram (Figure 2). Specific genes for each treatment were thus obtained, and their biological significance compared (Mapman functional category enrichment).
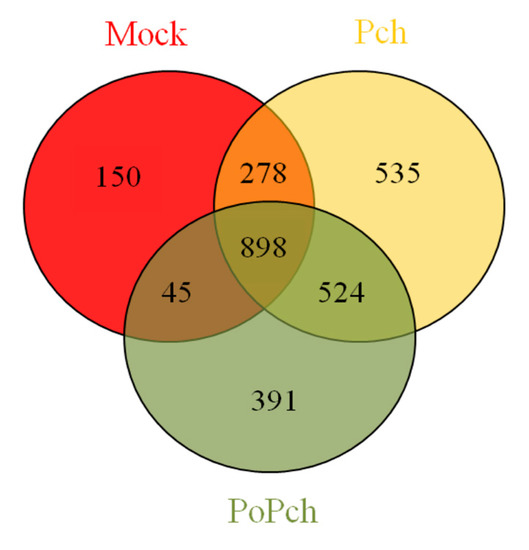
Figure 2. Venn diagram showing gene numbers in grapevine wood tissues common or specific to the three conditions considered ((i) mock inoculation, (ii) Phaeomoniella chlamydospora-wood infection, (iii) Phaeomoniella chlamydospora-wood infection and Pythium oligandrum Po2-root inoculation). Mock: mock inoculation; Po: P. oligandrum; Pch: P. chlamydospora; PoPch: P. oligandrum and P. chlamydospora.
The combined effect of all treatments revealed numerous DE genes (898), and analysis by functional categories using Mapman software indicated that the amplitude of expression appeared to play a major role. Although many interesting features can be obtained from these microarray result analyses, it is important to focus on the optimal points in high-throughput data regarding the biological system under investigation. In our case, these concerned (1) V. vinifera defense induction by P. chlamydospora and (2) V. vinifera induced resistance by P. oligandrum. The expected differences among treatments at the induction level for the most important DE genes, and statistically significant functional categories related to induced systemic resistance, ISR, (hormones, secondary metabolites, transcription factors and stress related genes), were effectively found. These results were observed when the genes were considered as common to all treatments, rather than when they were considered as specific to each treatment. Our analysis compared the expression of genes at 0-dpi against those at 14-dpi. Overexpression, for the most important functional categories involved in the establishment of ISR, took place at 0-dpi.
In all conditions where wounding occurred (mock inoculation and P. chlamydospora infection), the functional category presenting the strongest statistical significance was “secondary metabolism”. More detailed analyses revealed that several genes involved in phenylpropanoid biosynthesis (Figure 3) presented overexpression at 0-dpi (green genes) in the different treatments. These genes included four phenyl-alanine-ammonia lyase genes (PAL), one gene for 4-coumarate-CoA ligase (4CL) and three cinnamyl alcohol dehydrogenase (CAD) genes. One of the CAD genes was highly overexpressed at 0-dpi in plants treated with both microorganisms.
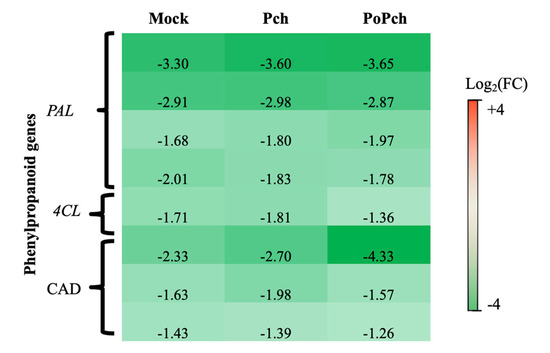
Figure 3. Characteristics of the grapevine transcriptome evolution for genes involved in the phenylpropanoid biosynthesis pathway between 0- and 14-dpi. Expression level values of phenylpropanoid genes (PAL: phenyl-alanine-ammonia lyase, 4CL: 4-coumarate-CoA ligase and CAD: cinnamyl alcohol dehydrogenase) are given as the fold change on a log2 basis. The red color means overexpression at 14-dpi and the green color means overexpression at 0-dpi. Mock: mock inoculation; Po: Pythium oligandrum; Pch: Phaeomoniella chlamydospora; PoPch: P. oligandrum and P. chlamydospora.
The “hormone” functional category was statistically significant for all treatments considered, with the jasmonate functional category having the largest number of DE genes (51) in the case of plants inoculated by both P. oligandrum and P. chlamydospora. Selected genes common to all treatments are presented in Figure 4. Two genes were particularly overexpressed at 0-dpi, but only in the joint presence of P. oligandrum and P. chlamydospora. One of the genes was involved in ET hormone regulation (log2 (FC) = −5.23), and the other one, annotated as allene oxide cyclase (log2(FC) = −6.12), was involved in the jasmonic acid (JA) biosynthesis pathway. Seven other genes of that pathway were induced at 0-dpi at various levels. One was lipoxygenase, two allene oxide synthase, and four related to 12 oxo-phytodienoic acid reductase, thereby confirming the strong involvement of the jasmonate pathway in the P. oligandrum induction of plant defenses.
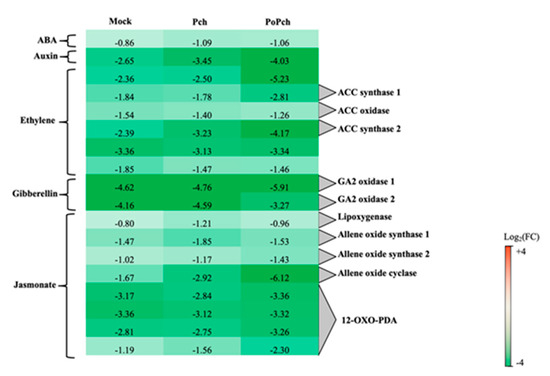
Figure 4. Characteristics of the grapevine transcriptome evolution for genes involved in hormone biosynthesis pathways between 0- and 14-dpi. Expression level values of genes encoding proteins involved in the synthesis or regulation of hormones (ABA: abscisic acid, ACC: 1-aminocyclopropane 1-carboxylate and GA: gibberellin) are given as the fold change on a log2 basis. The red color means overexpression at 14-dpi and the green color means overexpression at 0-dpi. Mock: mock inoculation; Po: Pythium oligandrum; Pch: Phaeomoniella chlamydospora; PoPch: P. oligandrum and P. chlamydospora.
Two other functional categories, stress-related genes and transcription factors, presented the induction of numerous genes in all treatments at 0-dpi. In the case of stress-related genes induction of abiotic- (heat shock proteins, wounding) and biotic-related genes (pathogenesis-related proteins and receptors), slight differences were detected according to the treatment. However, for transcription factors (Figure 5), P. oligandrum + P. chlamydospora treated plants showed globally a higher gene induction at 0-dpi than those inoculated with P. chlamydospora. Numerous functional categories were presented such as the Apetala2-Ethylene responsive element-binding protein (AP2-EREBP), Basic Helix-Loop-Helix (bHLH), MYB domain and WRKY domain. Moreover, AP2-EREBP was the most represented family, with several genes showing a stronger induction in the presence of P. oligandrum and P. chlamydospora.
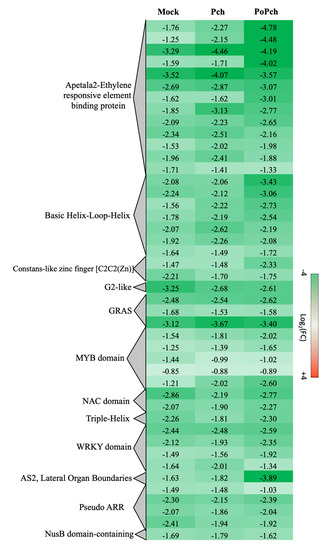
Figure 5. Global aspects of the grapevine transcriptome evolution for the transcription factor functional category between 0- and 14-dpi. Expression level values are given as a fold change on a log2 basis. The red color means overexpression at 14-dpi and the green color means overexpression at 0-dpi. Mock: mock inoculation; Po: Pythium oligandrum; Pch: Phaeomoniella chlamydospora; PoPch: P. oligandrum and P. chlamydospora.
References
- Gramaje, D.; Úrbez-Torres, J.R.; Sosnowski, M.R. Managing grapevine trunk diseases with respect to etiology and epidemiology: Current strategies and future prospects. Plant Dis. 2018, 102, 12–39.
- Mondello, V.; Songy, A.; Battiston, E.; Pinto, C.; Coppin, C.; Trotel-Aziz, P.; Clément, C.; Mugnai, L.; Fontaine, F. Grapevine trunk diseases: A review of fifteen years of trials for their control with chemicals and biocontrol agents. Plant Dis. 2018, 102, 1189–1217.
- Bertsch, C.; Ramírez-Suero, M.; Magnin-Robert, M.; Larignon, P.; Chong, J.; Abou-Mansour, E.; Spagnolo, A.; Clément, C.; Fontaine, F. Grapevine trunk diseases: Complex and still poorly understood. Plant Pathol. 2013, 62, 243–265.
- Kaplan, J.; Travadon, R.; Cooper, M.; Hillis, V.; Lubell, M.; Baumgartner, K. Identifying economic hurdles to early adoption of preventative practices: The case of trunk diseases in California winegrape vineyards. Wine Econ. Policy 2016, 5, 127–141.
- Calzarano, F.; Marco, S.D.; Cesari, A. Benefit of fungicide treatment after trunk renewal of vines with different types of esca necrosis. Phytopathol. Mediterr. 2004, 43, 10.
- Lorrain, B.; Ky, I.; Pasquier, G.; Jourdes, M.; Dubrana, L.G.; Gény, L.; Rey, P.; Donèche, B.; Teissedre, P.-L. Effect of Esca disease on the phenolic and sensory attributes of Cabernet Sauvignon grapes, musts and wines: Esca effect on grapes and wines composition. Aust. J. Grape Wine Res. 2012, 18, 64–72.
- Les Maladies du Bois de la Vigne—Réunion du Groupe de Travail Maladies du Bois. Available online: https://www.maladie-du-bois-vigne.fr/Zoom-sur/Reunion-du-Groupe-de-travail-Maladies-du-bois (accessed on 30 June 2020).
- Compant, S.; Brader, G.; Muzammil, S.; Sessitsch, A.; Lebrihi, A.; Mathieu, F. Use of beneficial bacteria and their secondary metabolites to control grapevine pathogen diseases. BioControl 2013, 58, 435–455.
- Mounier, E.; Boulisset, F.; Elbaz, N.; Dubournet, P.; Pajot, E. Esquive® WP limits development of grapevine wood diseases and reductions in the productive potential of land. In Proceedings of the 5th Conférence Internationale sur les Méthodes Alternatives de Protection des Plantes, Nouceau Sièle, Lille, France, 11–13 March 2015; Association Française de Protection des Plantes (AFPP): Alfortville, France, 2015; pp. 251–261.
- Blundell, R.; Lynch, M.; Haden, T.; Arreguin, M.; Gallagher, T.; Eskalen, A. Final Report: Evaluation of Vintec (Trichoderma atroviride_SC1) as Pruning Wound Protectants Against Selected Fungi Associated with Grapevine Trunk Diseases; Department of Plant Pathology, University of California: Davis, CA, USA, 2020.
- Gerbore, J.; Vallance, J.; Yacoub, A.; Delmotte, F.; Grizard, D.; Regnault-Roger, C.; Rey, P. Characterization of Pythium oligandrum populations that colonize the rhizosphere of vines from the Bordeaux region. FEMS Microbiol. Ecol. 2014, 90, 153–167.
- Benhamou, N.; le Floch, G.; Vallance, J.; Gerbore, J.; Grizard, D.; Rey, P. Pythium oligandrum: An example of opportunistic success. Microbiology 2012, 158, 2679–2694.
- Gerbore, J.; Benhamou, N.; Vallance, J.; Le Floch, G.; Grizard, D.; Regnault-Roger, C.; Rey, P. Biological control of plant pathogens: Advantages and limitations seen through the case study of Pythium oligandrum. Environ. Sci. Pollut. Res. 2014, 21, 4847–4860.
- Mohamed, N.; Lherminier, J.; Farmer, M.-J.; Fromentin, J.; Béno, N.; Houot, V.; Milat, M.-L.; Blein, J.-P. Defense responses in grapevine leaves against Botrytis cinerea induced by application of a Pythium oligandrum strain or its elicitin, oligandrin, to roots. Phytopathology 2007, 97, 611–620.
- Laveau, C.; Letouze, A.; Louvet, G.; Bastien, S.; Guérin-Dubrana, L. Differential aggressiveness of fungi implicated in esca and associated diseases of grapevine in France. Phytopathol. Mediterr. 2009, 48, 15.
- Daraignes, L.; Gerbore, J.; Yacoub, A.; Dubois, L.; Romand, C.; Zekri, O.; Roudet, J.; Chambon, P.; Fermaud, M. Efficacy of P. oligandrum affected by its association with bacterial BCAs and rootstock effect in controlling grapevine trunk diseases. Biol. Control 2018, 119, 59–67.
- Yacoub, A.; Gerbore, J.; Magnin, N.; Chambon, P.; Dufour, M.-C.; Corio-Costet, M.-F.; Guyoneaud, R.; Rey, P. Ability of Pythium oligandrum strains to protect Vitis vinifera L., by inducing plant resistance against Phaeomoniella chlamydospora, a pathogen involved in Esca, a grapevine trunk disease. Biol. Control 2016, 92, 7–16.
- Antonielli, L.; Compant, S.; Strauss, J.; Sessitsch, A.; Berger, H. Draft genome sequence of Phaeomoniella chlamydospora strain RR-HG1, a grapevine trunk disease (esca)-related member of the ascomycota. Genome Announc. 2014, 2, e00098-14.
- Blanco-Ulate, B.; Rolshausen, P.; Cantu, D. Draft genome sequence of the Ascomycete Phaeoacremonium aleophilum strain UCR-PA7, a causal agent of the esca disease complex in grapevines. Genome Announc. 2013, 1, e00390-13.
- Robert-Siegwald, G.; Vallet, J.; Abou-Mansour, E.; Xu, J.; Rey, P.; Bertsch, C.; Rego, C.; Larignon, P.; Fontaine, F.; Lebrun, M.-H. Draft genome sequence of Diplodia seriata F98.1, a fungal species involved in grapevine trunk diseases. Genome Announc. 2017, 5, e00061-17.
References
- Gramaje, D.; Úrbez-Torres, J.R.; Sosnowski, M.R. Managing grapevine trunk diseases with respect to etiology and epidemiology: Current strategies and future prospects. Plant Dis. 2018, 102, 12–39.
- Mondello, V.; Songy, A.; Battiston, E.; Pinto, C.; Coppin, C.; Trotel-Aziz, P.; Clément, C.; Mugnai, L.; Fontaine, F. Grapevine trunk diseases: A review of fifteen years of trials for their control with chemicals and biocontrol agents. Plant Dis. 2018, 102, 1189–1217.
- Bertsch, C.; Ramírez-Suero, M.; Magnin-Robert, M.; Larignon, P.; Chong, J.; Abou-Mansour, E.; Spagnolo, A.; Clément, C.; Fontaine, F. Grapevine trunk diseases: Complex and still poorly understood. Plant Pathol. 2013, 62, 243–265.
- Kaplan, J.; Travadon, R.; Cooper, M.; Hillis, V.; Lubell, M.; Baumgartner, K. Identifying economic hurdles to early adoption of preventative practices: The case of trunk diseases in California winegrape vineyards. Wine Econ. Policy 2016, 5, 127–141.
- Calzarano, F.; Marco, S.D.; Cesari, A. Benefit of fungicide treatment after trunk renewal of vines with different types of esca necrosis. Phytopathol. Mediterr. 2004, 43, 10.
- Lorrain, B.; Ky, I.; Pasquier, G.; Jourdes, M.; Dubrana, L.G.; Gény, L.; Rey, P.; Donèche, B.; Teissedre, P.-L. Effect of Esca disease on the phenolic and sensory attributes of Cabernet Sauvignon grapes, musts and wines: Esca effect on grapes and wines composition. Aust. J. Grape Wine Res. 2012, 18, 64–72.
- Les Maladies du Bois de la Vigne—Réunion du Groupe de Travail Maladies du Bois. Available online: https://www.maladie-du-bois-vigne.fr/Zoom-sur/Reunion-du-Groupe-de-travail-Maladies-du-bois (accessed on 30 June 2020).
- Compant, S.; Brader, G.; Muzammil, S.; Sessitsch, A.; Lebrihi, A.; Mathieu, F. Use of beneficial bacteria and their secondary metabolites to control grapevine pathogen diseases. BioControl 2013, 58, 435–455.
- Mounier, E.; Boulisset, F.; Elbaz, N.; Dubournet, P.; Pajot, E. Esquive® WP limits development of grapevine wood diseases and reductions in the productive potential of land. In Proceedings of the 5th Conférence Internationale sur les Méthodes Alternatives de Protection des Plantes, Nouceau Sièle, Lille, France, 11–13 March 2015; Association Française de Protection des Plantes (AFPP): Alfortville, France, 2015; pp. 251–261.
- Blundell, R.; Lynch, M.; Haden, T.; Arreguin, M.; Gallagher, T.; Eskalen, A. Final Report: Evaluation of Vintec (Trichoderma atroviride_SC1) as Pruning Wound Protectants Against Selected Fungi Associated with Grapevine Trunk Diseases; Department of Plant Pathology, University of California: Davis, CA, USA, 2020.
- Gerbore, J.; Vallance, J.; Yacoub, A.; Delmotte, F.; Grizard, D.; Regnault-Roger, C.; Rey, P. Characterization of Pythium oligandrum populations that colonize the rhizosphere of vines from the Bordeaux region. FEMS Microbiol. Ecol. 2014, 90, 153–167.
- Benhamou, N.; le Floch, G.; Vallance, J.; Gerbore, J.; Grizard, D.; Rey, P. Pythium oligandrum: An example of opportunistic success. Microbiology 2012, 158, 2679–2694.
- Gerbore, J.; Benhamou, N.; Vallance, J.; Le Floch, G.; Grizard, D.; Regnault-Roger, C.; Rey, P. Biological control of plant pathogens: Advantages and limitations seen through the case study of Pythium oligandrum. Environ. Sci. Pollut. Res. 2014, 21, 4847–4860.
- Mohamed, N.; Lherminier, J.; Farmer, M.-J.; Fromentin, J.; Béno, N.; Houot, V.; Milat, M.-L.; Blein, J.-P. Defense responses in grapevine leaves against Botrytis cinerea induced by application of a Pythium oligandrum strain or its elicitin, oligandrin, to roots. Phytopathology 2007, 97, 611–620.
- Laveau, C.; Letouze, A.; Louvet, G.; Bastien, S.; Guérin-Dubrana, L. Differential aggressiveness of fungi implicated in esca and associated diseases of grapevine in France. Phytopathol. Mediterr. 2009, 48, 15.
- Daraignes, L.; Gerbore, J.; Yacoub, A.; Dubois, L.; Romand, C.; Zekri, O.; Roudet, J.; Chambon, P.; Fermaud, M. Efficacy of P. oligandrum affected by its association with bacterial BCAs and rootstock effect in controlling grapevine trunk diseases. Biol. Control 2018, 119, 59–67.
- Yacoub, A.; Gerbore, J.; Magnin, N.; Chambon, P.; Dufour, M.-C.; Corio-Costet, M.-F.; Guyoneaud, R.; Rey, P. Ability of Pythium oligandrum strains to protect Vitis vinifera L., by inducing plant resistance against Phaeomoniella chlamydospora, a pathogen involved in Esca, a grapevine trunk disease. Biol. Control 2016, 92, 7–16.
- Antonielli, L.; Compant, S.; Strauss, J.; Sessitsch, A.; Berger, H. Draft genome sequence of Phaeomoniella chlamydospora strain RR-HG1, a grapevine trunk disease (esca)-related member of the ascomycota. Genome Announc. 2014, 2, e00098-14.
- Blanco-Ulate, B.; Rolshausen, P.; Cantu, D. Draft genome sequence of the Ascomycete Phaeoacremonium aleophilum strain UCR-PA7, a causal agent of the esca disease complex in grapevines. Genome Announc. 2013, 1, e00390-13.
- Robert-Siegwald, G.; Vallet, J.; Abou-Mansour, E.; Xu, J.; Rey, P.; Bertsch, C.; Rego, C.; Larignon, P.; Fontaine, F.; Lebrun, M.-H. Draft genome sequence of Diplodia seriata F98.1, a fungal species involved in grapevine trunk diseases. Genome Announc. 2017, 5, e00061-17.
