Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Deog-Hwan Oh and Version 2 by Vivi Li.
The prevalence of metabolic syndrome (MetS) is presently an alarming public health problem globally. Oxidative stress has been postulated to be strongly correlated with MetS, such as type 2 diabetes, obesity, hypertension, cardiovascular diseases, and certain cancers. Cereals are important staple foods which account for a huge proportion of the human diet. However, owing to recent growing demand and the search for natural antioxidants for the prevention and management of MetS, cereal peptides have gained increasing attention for developing functional ingredients or foods with substantial antioxidant properties.
- Cereal grains
- Antioxidant peptides
- Oxidative stress
- Metabolic syndrome
- Functional food
1. Oxidative Stress
The term “oxidative stress” was initially conceptualized about three (3) decades ago [1][2][1,2]. Since then, the term has evolved remarkably. The inception of oxidative stress finds its roots in early publications of Seyle (loc. cit), which were concerned with the toxicity of oxygen linked with aging, bodily responses and processes associated with oxygen radicals, the concept of the physiology of the mitochondria and research on its aging, as well as work on variances of redox reactions in living organisms [1]. Table 1 shows several definitions proposed by different scientists over the years. Consequently, oxidative stress is denoted by an imbalance between oxidant and antioxidant species, such that oxidant species weigh more, which results in the excessive release of free radicals or reactive oxygen species (ROS) and causes cellular and molecular disruption, as well as a negative influence on redox signaling [3][4][5][3,4,5]. Oxidative stress occurs when antioxidant defenses are impaired or are not strong enough to overpower the production of reactive oxygen species (ROS) [3][4][6][3,4,6]. The consequences of oxidative stress may be progressive and often dire. The two underlying components in oxidative stress, as evidenced by the definition above, are prooxidant and antioxidant species. It is their non-homeostatic co-existence that results in oxidative stress. To prevent excess ROS production in mammalian cells, antioxidant molecules and antioxidant enzymes act as defense systems. There are many prooxidant species and antioxidant species. In cells, glutathione (GSH) is the most abundant and important non-protein antioxidant molecule. Antioxidants are those substances that counteract the harmful effects of oxidants. They are usually produced in insufficient quantities by the body; therefore, they need to be supplemented frequently from external sources, typically food sources [7]. Examples of antioxidants are vitamins A, C, E, carotenoids (including beta-carotene and astaxanthins), and polyphenols (such as flavonoids, isoflavones, anthocyanins, chlorogenic, and catechins) [7][8][9][7,8,9]. Examples of prooxidants, particularly the reactive species, include reactive oxygen species (ROS) [6][10][6,10], reactive sulfur species (RSS) [11], reactive electrophile species (RES) [12], reactive carbonyl species (RCS) [13][14][13,14], reactive nitrogen species (RNS) [15][16][17][15,16,17], and reactive halogen species (RHS) [10][15][18][10,15,18]. ROS, RHS, and RNS are toxic oxidants that cause damage to DNA, RNA, lipids, and phagocytosed pathogen proteins, especially during inflammation [10]. According to Yang et al. [19], ROS, RHS, and RNS cause apoptosis by directly oxidizing protein, lipid, and DNA signaling pathways, which increases the risk of CVD, specifically atherosclerosis. RSS are molecules produced from sequential one-electron oxidations (loss of electrons in a chemical reaction) of hydrogen sulfide, thus forming thiyl, hydrogen persulfide, and the persulfide “supersulfide” radicals, before terminating in elemental sulfur [11]. There are resemblances between ROS and RSS, and they are sometimes misconstrued and used interchangeably. However, RSS have more effectiveness, reactivity, signaling, and versatility potential compared to ROS. They can also be accumulated and reused [11]. RES have a wide range of functionality with overlying chemical reactivity, which sometimes makes the study of biological RES challenging. Biologically, they range in different shapes and forms. RES involved in cell signaling may even rise higher when they sense the human body is stressed. RHS control antioxidant response, cell growth, DNA damage development, aging, cellular homeostasis events, such as apoptosis, and immune response [12].
Table 1.
Some proposed definitions of oxidative stress throughout the course of time.
| Definition of Oxidative Stress | Reference |
|---|---|
| Oxidative stress is a disturbance in the prooxidant–antioxidant balance in favor of the former. | [20][24] |
| Oxidative stress is defined as a disturbance in the prooxidant–antioxidant balance that leads to potential damage. | [21][25] |
| Oxidative stress is a situation when steady-state ROS concentration is transiently or chronically enhanced, disturbing cellular metabolism, its regulation, and damaging cellular constituents. | [22][20] |
| Oxidative stress is defined as excess production of reactive oxygen species (ROS) relative to antioxidant defense. | [23][26] |
| Oxidative stress refers to the imbalance between cellular antioxidant cascade and processes that generate reactive oxygen species (ROS), such as superoxide (O2.), hydrogen peroxide (H2O2), and hydroxyl anion (OH−). | [24][27] |
| Oxidative stress is defined as an imbalance between oxidants and antioxidants in favor of the oxidants, leading to a disruption of redox signaling and control and/or molecular damage. This implies a deviation to the opposite side of the balance (thus, “reductive stress”), as well as physiological and supraphysiological deviations, which tie into the overarching concept of “oxidative distress” and “oxidative eustress”, respectively. | [5] |
The oxidation of macromolecules, such as carbohydrates, lipids, and amino acids, produces many reactive carbonyl species (RCS). RCS can react and cause changes to the surface composition of proteins, nucleic acids and amino phospholipids, and therefore serve as agents of cell destruction and gene mutation. Furthermore, interaction of RCS with biological samples results in many chemical products that have diverse negative effects on human health [13]. RCS are shown to be involved in ROS signaling, and are typically classified under RES [14]. Electrophilic and nucleophilic reactions are some of the most common covalent bond formations that are found in many chemical–cellular reactions. The electrophilic nature of carbonyl compounds has a high affinity for nucleophilic cellular constituents, thus making it possible for easy accessibility of the cells by RCS to render its physiological effects [13][14][13,14]. In the form of free radicals, such as nitric oxide and other nitric oxide-derived species (organic species, such as 3-nitrotyrosine (3-NT) and S-nitrosothiols (RS–NO), and inorganic species, such as nitrite [15]), RNS are involved in a variety of biological activities, and their detection and quantification are technically difficult [15][16][15,16]. However, there is evidence that, when RNS is produced in excess, it disrupts protein synthesis in the body, which harms mitochondrial metabolism dynamics and mitophagy in the nervous system [17]. RHS, especially those with chlorine, bromine, and iodine (thus, HOX with X=Cl, Br or I) induce injuries to DNA, RNA, lipids, and proteins cells [10][15][10,15]. They render the immune system’s defense useless when they are released into the body in high quantities by reversing the action of phagocytes (including neutrophils, monocytes, macrophages, mast cells, dendritic cells, osteoclasts, and eosinophils) that capture pathogens, and, thus, instigate disease or infection pathogenesis [10]. When RHS interreact with extracellular myeloperoxidase (MPO) produced by neutrophils to provide immune system defenses against pathogens, it results in tissue damage, cellular damage, and inflammatory reactions that facilitate the occurrence of a variety of diseases, particularly CVD (atherosclerosis), obesity, T2DM, and, ultimately, metabolic syndrome (MetS) [10][18][10,18]. Owing to the vast quantities and differences in prooxidant and antioxidant species, the best way to understand oxidative stress is through a classification system. According to Sies et al. [4], oxidative stress can be classified according to its intensity (basal, low, intermediate, and high), specific forms, related terms, and associated biological responses (Figure 1). A further assessment by Lushchak [22][20] establishes that imbalance could also result from one or a combination of the following: elevated levels of endogenous and exogenous compounds entering autoxidation, coupled with ROS production; depleted stores of low molecular mass antioxidants; deactivated antioxidant enzymes; and reduced production of antioxidant enzymes and low molecular mass antioxidants. These determine the three types of oxidative stress—acute, chronic, and quasi-stationery [22][25][26][27][20,21,22,23].
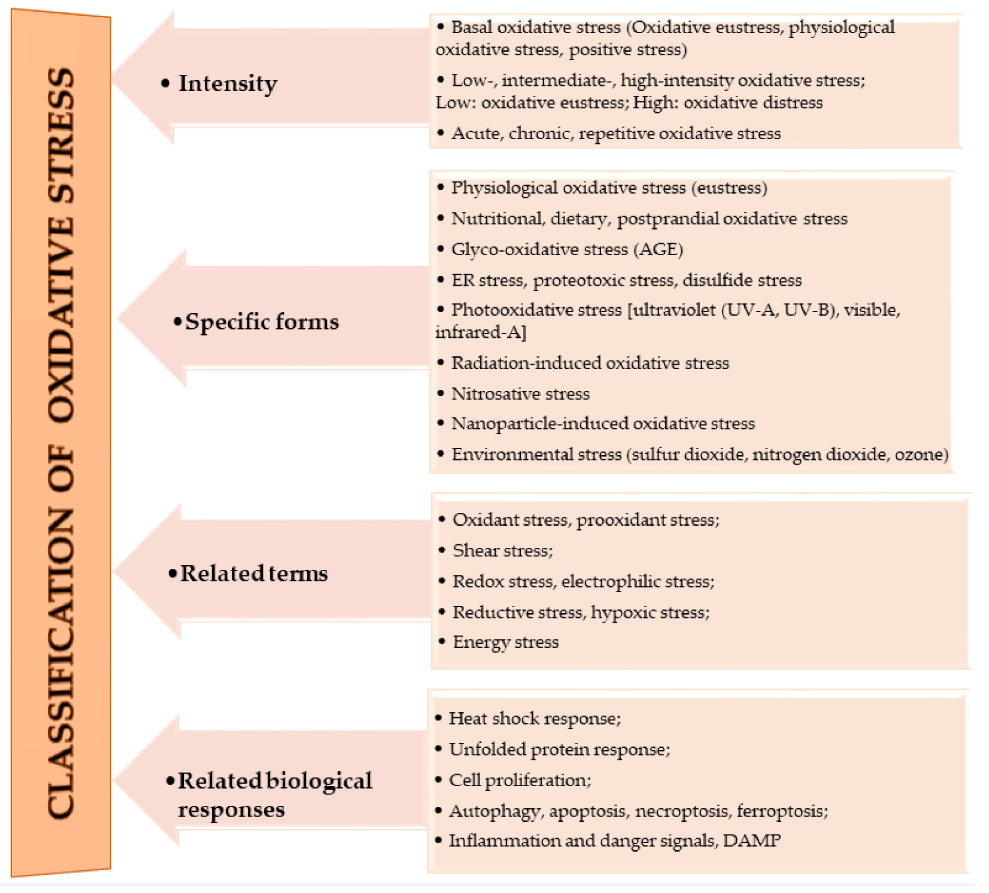
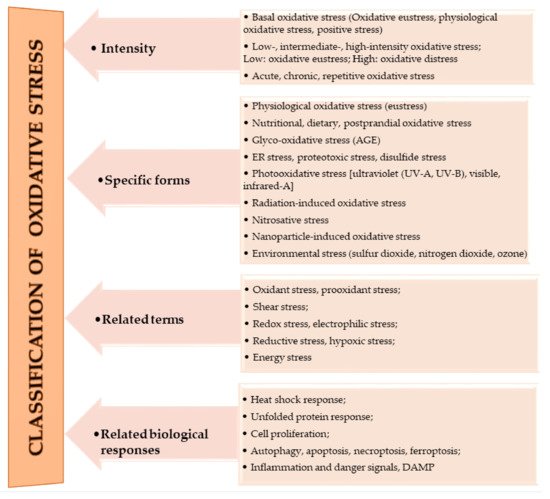
Figure 1.
Classifications of oxidative stress adapted with permission from Sies et al. [4]. Copyright 2017 Annual Reviews, Inc.
2. Production Strategies for Antioxidant Peptides
Antioxidant peptides, like other functional peptides, are encrypted within the sequence of parent protein molecules and can be liberated through proteolytic enzyme hydrolysis, gastrointestinal digestion, or microbial fermentation. However, antioxidant peptides have been produced largely using enzymatic hydrolysis. In recent times, innovative and efficient food processing techniques, such as high hydrostatic pressure (HHP), microwave assistance (MA), ultrasonic assistance, pulsed electric field (PEF), and subcritical water have been employed in combination, or simultaneously, with conventional (classical) methods to improve yields and produce mono- or multifunctional peptides with lesser cost and time. These emerging technologies have been comprehensively discussed in recent reviews [28][29][48,134]. The production of antioxidant peptides from cereals and other plant sources is shown in Figure 23, and involves protein isolate pretreatment with novel food processing technologies (HHP, MA, PEF etc), followed by protease hydrolysis to obtain protein hydrolysate. Further purification steps using membrane separation, chromatographic methods, including ion–exchange chromatography (IEC), gel permeation chromatography (GPC), and reversed–phase high performance liquid chromatography (RP-HPLC), are performed to obtain antioxidant peptides, which are later identified by mass spectrometry (electrospray and tandem) and their activity is evaluated in vitro and in vivo tests [30][31][66,135].
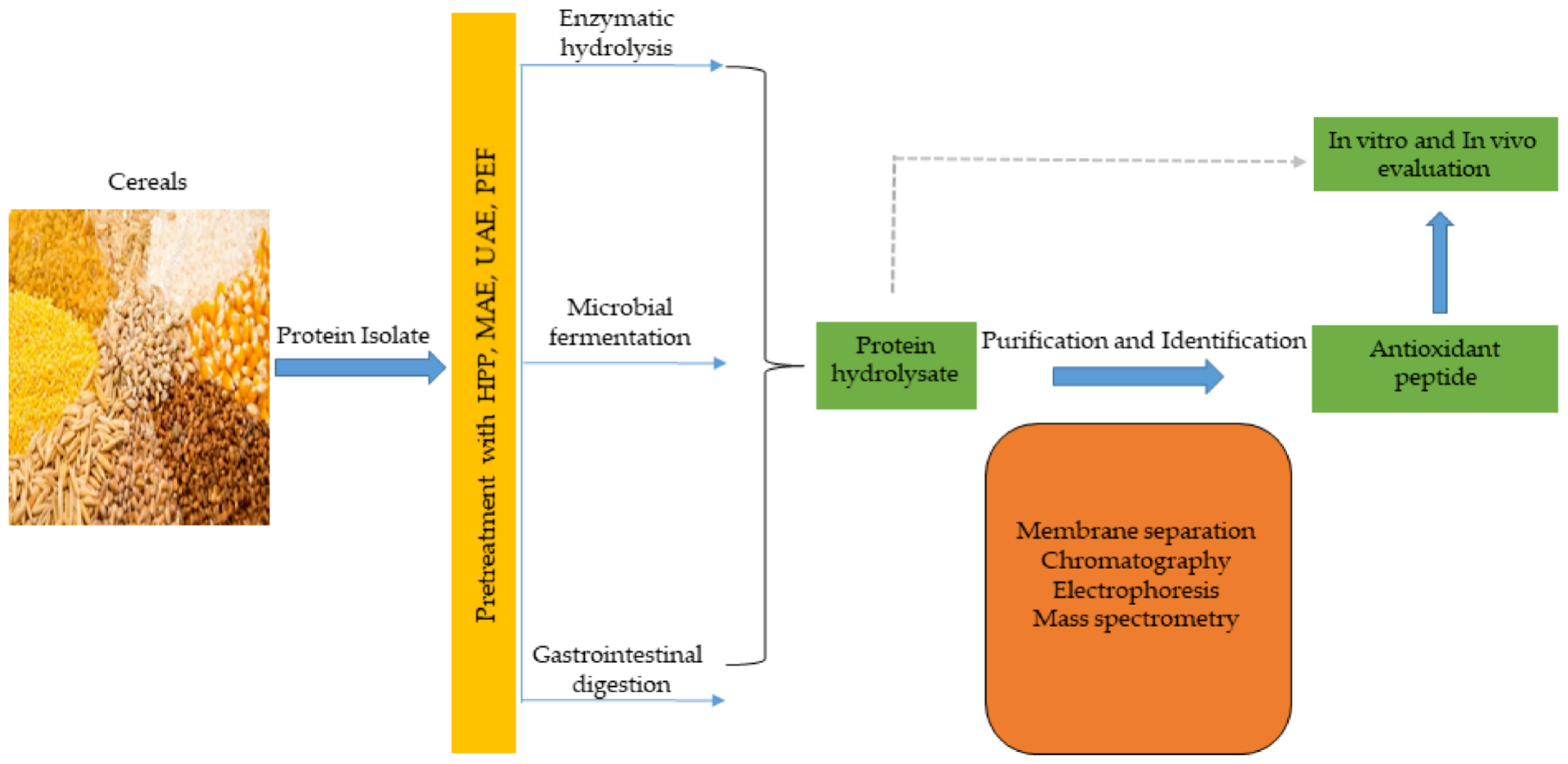
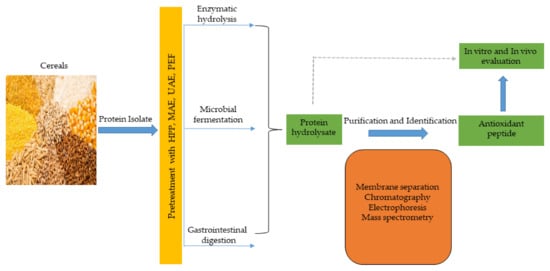
Figure 23.
Cereal peptidic antioxidants production strategies.
2.1. Conventional Approach
2.1.1. Enzymatic Hydrolysis
Hydrolysis of food proteins to produce multi- or monofunctional peptides has been mainly conducted using enzymes. Bioactive peptides have gained much attention over the years and there has recently been particular interest in antioxidative peptides due to their potential multifunctional activities in the prevention and management of several diseases. Exogenous enzymes, specifically those from microbial sources (Alcalase, Flavourzyme and ProtamexTM), animal sources (pepsin and trypsin), and plant sources (e.g., papain), have extensively been used to produce antioxidative peptides [32][136]. Compared with the preparation methods of other peptides, the enzymatic hydrolysis method has the advantages of absence of organic solvents or toxic chemicals, easy reaction control, good repeatability, low cost, and low energy consumption. Therefore, it is the most widely preferred method for industrial production. However, decreasing hydrolysis time, the amount of enzyme, and improving the yield and bioactivity of peptides remain main challenges to be circumvented [33][137]. Consequently, innovative and emerging technologies, such as high hydrostatic pressure (HHP), microwave, and pulsed electric field, are being used to induce protein unfolding and activation of enzymes, thereby improving the efficiency of enzyme hydrolysis and generating peptide fractions with strong antioxidant activities [29][134]. The application of emerging technologies to produce high value BP will be discussed in detail later in this rentryview.
The generation of protein hydrolysates and/or peptides with desirable functional properties is mainly affected by several factors, such as the types of enzyme and hydrolysis conditions (temperature, pH, the ratio of enzyme to substrate, and time), among others. It is worth noting that different types of enzymes can specifically cleave different positions of the peptide. For example, pepsin digestion increases hydrophobicity by cleaving peptide bonds between hydrophobic amino acids (preferably aromatic amino acids, including Phe, Trp, and Tyr), resulting in an appropriate reaction of rice bran protein hydrolysates (RBPH) with DPPH free radicals. Nonetheless, a reduction in this ability was observed due to the action of trypsin, which further selectively cleaves basic amino acids [34][120]. Thus, the type of peptidase is crucial in determining the size, composition, and amino acid sequence of the peptides, and thus affects antioxidant activity [35][138]. Peptidase is mainly divided into animal, plant, and microbial proteases. In general, pepsin and trypsin are the most widely used animal proteases. Suetsuna and Chen [36][111] reported that the two peptides (Leu-Gln-Pro-Gly-Gln-Gly-Gln-Gln-Gly and Ala-Gln-Ile-Pro-Gln-Gln) obtained from wheat gluten protein by using pepsin have good antioxidant capacities. The digestion of food proteins by digestive enzymes, or enzymes produced by microbial flora residing in the gut, offers another opportunity to produce peptides with antioxidative activities directly or indirectly in the gut, or through cell signaling pathways or their ability to enter blood circulation by crossing epithelial cell membrane to target sites [32][37][136,139]. Therefore, the stability or resistance of food-derived antioxidative peptides to GI breakdown or modification is an essential factor to be considered to ensure they exert their beneficial health properties.
2.1.2. Microbial Fermentation
Microorganisms degrade food components through their complex enzyme systems. Bacteria belonging to the Lactobacillus genus are largely used for microbial fermentation to produce foods with improved nutritional properties and functional health benefits. Lactic acid bacteria (LAB) have a long history of use in fermented foods, and are generally regarded as safe (GRAS) [38][39][140,141]. An extracellular proteinase, a transport system specific for small peptides, and a multitude of intracellular specific, generic, endo-, and eso-peptidases constitute the proteolytic system of LAB [40][142]. Cereals are key components of daily diets in most parts of the world and, hence, their fermentation generates peptides with great biological importance. Antioxidant properties of cereals have been improved via the use of microbial bioprocessing and endogenous proteolytic enzymes to release antioxidant peptides and phenolic compounds from plant matrix. Antioxidant peptides have been produced and isolated from the sourdough fermentation of cereals [41][143]. Different strains have been reported to possess different proteolytic and peptidase activities on cereal proteins to release several peptides [39][41][42][141,143,144]. Galli et al. [41][143] revealed that different lactobacilli showed specific proteolytic and peptidase activity in wheat sourdough, which resulted in the production of low molecular weight peptides that exerted antioxidant and anti-inflammatory activities on RAW 264.7 murine macrophage, murine H-end endothelium cells, and human intestinal Caco-2 cells.
The effects of cooking on the anti-inflammatory and antioxidant properties of wheat sourdoughs and bread produced by three Lactobacilli strains (L. farciminis H3 and A11 and L. sanfranciscensis I4) were assessed by Luti et al. [43][145]. Peptides from dough and bread were found to suppress the NFkB pathway and, also, to reduce intracellular ROS levels. Biological activities were retained after cooking, despite differences in amino acid compositions and sequences between dough and bread peptides. Wheat germ fermented with Lactobacillus plantarum DY-1 exhibited a hydroxyl radical scavenging capacity of 72.8 ± 2.9% and retarded thiobarbituric acid-reactive substances (TBARSs) formation in emulsified sausages stored at 4 °C for 7 days [44][146]. Niu et al. [45][147] found promising antioxidant activities from peptides (less than 1 kDa) obtained by fermentation of wheat germ with Bacillus Subtilis B1. They observed significant EC50 dose-dependent DPPH, hydroxyl and superoxide anion radical scavenging activities of 3.16 mg/mL, 6.04 mg/mL and 7.46 mg/mL, respectively. Studies by Wang et al. [46][148] showed that co-fermentation of barley with Lactobacillus plantarum and Rhizopus oryzae increased amino acid nitrogen, <10 kDa peptide, and free phenolic contents, and thus improved DPPH, hydroxyl, ABTS+ radical scavenging activity, and ferric reducing antioxidant power. Hydrolysates obtained during the fermentation of amaranth protein fractions with Lactobacillus helveticus and Lactobacillus plantarum possessed higher peroxyl and hydroxyl radical scavenging activities [47][149].
2.2. Bioinformatics Approach
Owing to the challenges involved in developing BP using classical approaches, which include time consumption, high cost, and uncertainties regarding the bioactivities of protein hydrolysates or fragment peptides which needs to be validated, bioinformatics could be a promising tool to discover bioactive peptides from different protein sources [48][49][150,151]. Bioinformatics, or in silico analysis, employs computational and statistical techniques to manage, curate, predict and interpret biological datasets [48][150]. Bioinformatics can minimize the number of experiments that must be performed to prepare BP by determining how their structure relates to their activity. Recently, researchers have employed the in silico approach to predict the production of BP from food proteins using bioinformatics and databases. This strategy, combined with classical approaches, can determine the optimum BP production parameters, such as the type of enzyme and target activity. In silico analysis has been used to predict peptides released by single and multiple enzyme digestion. Amino acid sequences and positions are key determinants of the bioactivities of peptides [50][152]. Several studies have attributed the bioactivies of peptides to the presence of some specific amino acids. For example, the amino acids cysteine (C), histidine (H), proline (P), methionine (M), and aromatic amino acids of food peptides have been reported to exert antioxidant activity [51][153]. Molecular docking approaches are used to predict and estimate the binding modes and affinities of small molecules within the binding sites of target receptors [52][154]. They have been used to screen for food-derived BP and illustrate their biological mechanisms. Protein structure selection and preparation, ligand preparation, docking, and analysis of results are the main steps involved in molecular docking [49][151]. In addition, bioinformatics approaches could be used to simulate and predict gastrointestinal stability, toxicity and allergenicity of peptides. Nonetheless, in vitro and in vivo validation of such a prediction needs to be carried out and ascertained. The bioavailability of antioxidant peptides is mainly affected by their transepithelial transport, and the human Caco-2 cell model is widely used for in vitro studies to investigate potential relevance in in vivo metabolism [31][135]. The in vivo challenges encountered by peptides as therapeutics has been comprehensively discussed by Yap and Gan [53][155]. Despite its wide use, increasing prediction accuracy of this computational tool can help overcome some theoretical and computational drawbacks. Quantitative structure–activity relationship (QSAR) modeling reveals how the structural characteristics of compounds relates to their biochemical and functional properties. QSAR model development involves the following steps: (i) retrieving sequences of target peptides from a database or library; (ii) scalar description of amino acids constituents; (iii) QSAR model construction and activity prediction; and (iv) validation of synthesized peptides in vitro or in vivo. However, QSAR approaches are not without their limitations, as model development is difficult with lack of knowledge and the unavailability of protein sequences in protein libraries and online databases [54][156]. The reliability and predictability of a three-dimensional quantitative structure–activity relationship (3D-QSAR) model was developed using a combination of comparative molecular field analysis (CoMFA) and comparative similarity index analysis (CoMSIA), for a total of 198 antioxidant tripeptides retrieved from literature. Promising antioxidant activity was demonstrated from graphical contour maps of the model with significant contribution by electrostatic, steric, hydrophilic and hydrogen bond acceptor force fields. Consequently, ten novel tripeptides were designed with residue substitution, and their antioxidant activities were predicted by the model. Subsequently, the tripeptides were synthesized and validated by FRAP (ferric reducing antioxidant Potential) and ABTS (2,2′-azino-bis (3-ethlbenzthiazoline-6-sulfonic acid)) assays. Tripeptides WKW, GRC, ARW, LRW, LKW, and YKW showed higher ferric reducing capacity and ABTS radical scavenging capacity. Findings from this work showed a high correlation between experimental and predicted activity, and the developed model could provide insight regarding the structure and activity relationship of antioxidant peptides and be useful in their virtual screening and design [55][157]. Yan et al. [56][158] designed two novel tripeptides, GWY and QWY, using 3D-QSAR models, which demonstrated strong antioxidant activities of 3.32 mM TE and 2.97 mM TE, respectively; after synthesis and in vitro confirmatory evaluation using Trolox equivalent antioxidant capacity (TEAC) assay. These authors further investigated the potential molecular mechanism using molecular docking and molecular dynamics simulations. Their findings revealed that GWY and QWY could improve the body’s antioxidant defense system by competitively binding to Keap1′s active sites key residues Arg415, Arg483, Arg380 and Ser555, increasing the accumulation of Nrf2 and, hence, activating the Kelch-like ECH associated protein1 (Keap1)-nuclear factor erythroid 2-related factor 2 (Nrf2)-antioxidant response element (ARE) signaling pathway.2.3. Emerging Food Processing Technologies
The potential application of innovative and emerging food processing techniques to improve food-derived protein digestibility, and produce BP of interest, is increasingly being explored (Table 25). HHP, a non-thermal and green technology which instantaneously and uniformly transmits isostatic pressure (100–1000 MPa) to enhance shelf life, improves the functional and bioactive properties of food products, and is an effective strategy to produce antioxidant peptides from various food sources. HHP treatment induces denaturation of native protein by disrupting hydrogen, as well as hydrophobic and ionic bonds, but not covalent and non-covalent bonds; hence, it modifies protein secondary structure but not the primary structure. Without the use of high temperatures, pressurization may improve the susceptibility of unfolded proteins access to enzyme hydrolysis [57][58][159,160]. Ultrasonication uses microbubble cavitation, and is considered an environmentally friendly food processing technique with higher yield, extraction rate, reproducibility, purity, minimal energy, water and solvent use. However, several factors, including ultrasound power, intensity, frequency, temperature, solvent, reactor design, as well as matrix parameters, are known to beneficially or negatively influence food components and metabolites [59][161]. Ultrasonication has been shown to increase protein extraction and sorghum digestibility. Sullivan et al. [60][162] found that ultrasonication at 40% amplitude for 10 min increased the solubility of sorghum kafirin protein from 6.5 μg/mL to 173.3 μg/mL, as well as its digestibility as a result of its secondary structure modification. In addition, ultrasonication followed by in vitro pepsin–pancreatin hydrolysis improved the antioxidant capacity of purified kafirin and sorghum gluten-like flour. Thus, ultrasonication could serve as a potential technique to improve the nutritional benefits and functionality of sorghum flour. Electron beam irradiation (EBI), an ionizing irradiation, is a safe, nonthermal, less expensive, and environmentally friendly technique used widely to modify food components and improve functional properties [61][163]. The functional and antioxidant properties of alcalase hydrolysates of wheat germ protein was remarkably improved after EBI treatment [62][164]. Li et al. [63][165] assessed the effect of EBI treatment on the structure and antioxidant activity of rice protein after alcalase hydrolysis. Even though EBI treatment induced amino acid oxidation as irradiation doses increased to 50 kGy, it caused changes in the secondary structure and hydrophobic regions in protein cores, leading to more fragmentations of hydrolysates and improvement in the antioxidant ability of rice protein. Irradiation at 50 kGy increased the DPPH and ABTS radical scavenging activity to 96.8% and 92.0%, respectively, compared to the 66.7% and 71.1% observed in non-irradiated rice protein hydrolysates.Table 2. Cereal antioxidant peptides using different processing methods.
| Protein Source | Processing Method | Peptide Sequence/Size/Hydrolysate |
Assay Outcome | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wheat germ protein hydrolysates | Electron beam irradiation (EBI) | <1 kDa | Irradiation at 50 kGy increased DPPH and ABTS+ radical scavenging activity by 45.77% and 52.52%, respectively. | [ | ||||||
| Wheat germ | 62 | ] | ||||||||
| Trypsin, Alcalase | AREGETVVPG | Vascular smooth muscle cells | High glucose-induced cell growth and generation of intracellular ROS was significantly decreased by AOP (5 µM). Suppression of PKCζ, AKT and Erk1/2 phosphorylation, and inhibition of Nox4 protein expression by AOP (5 µM). | [72] | Wheat germ | Fermentation with Bacillus subtilis | N.R | Peptide content increased in fermented samples compared to non-fermented samples and resulted in an increase in DPPH radical scavenging, Fe2+ chelating and Fe3+ reducing power activities. | [64] | |
| Wheat germ | neutral protease | ADWGGPLPH | Vascular smooth muscle cells | High glucose-induced cell proliferation and intracellular ROS generation was significantly reduced by peptide at 10 µM and 20 µM. Stimulation of AMPK activity, inhibition of PKCζ, AKT and Erk1/2 phosphorylation, and suppression of NOX4 protein expression. | [71] | KAMUT® Wheat | Combination of enzyme hydrolysis (Alcalase, Neutrase, Flavourzyme) and fermentation with (Lactobacillus spp. strains) | VLPPQQQY | Stronger superoxide anion, hydroxyl radicals, organic nitro-radicals (ABTS, DPPH) scavenging, and lipid peroxidation inhibition was observed. | [65] |
| Wheat gluten protein | Alcalase | Protein hydrolysate | Human peripheral blood mononuclear cells | Hydrolysate (0.5 mg/mL) directly scavenged free radicals, increased GSH levels, reduced NO overproduction, and, thus, enhanced cells’ antioxidant capacity. Also, cell proliferation, Th1 and Th17 pro-inflammatory cytokines IFN-γ and and IL-17 were reduced. | [73] | Wheat germ | Subcritical water extraction | GPFGPE, FGE, <1 kDa | Peptide fraction 4 showed the strongest DPPH radical scavenging activity and could effectively cross Caco-2 intestinal epithelium cells. | |
| Foxtail millet | Alcalase | PFLF, IALLIPF[66] | ||||||||
| Human keratinocyte HaCaT cells | ROS, MDA production was effectively reduced and GSH levels increased by MPP (300 µg/mL) in H | 2O2-induced HaCaT cells. | [74] | Sorghum | Ultrasonication combined with pepsin-pancreatin hydrolysis | <1 kDa | Ultrasonication increased DH, DPPH scavenging activity and ORAC values. However, there was no improvement in NO scavenging activity. | [60] | ||
| Rice dregs | Angling method using metal-organic framework combined with alcalase hydrolysis. | GDMNP, LLLRW | Strong DPPH, superoxide anion, hydroxyl radical scavenging and Fe | |||||||
| Sorghum | Pepsin-pancreatin | Kafirin hydrolysate | THP-1 human macrophages | Kafirin (100 μg/mL) reduced LPS-induced intracellular ROS production, inflammatory cytokines (IL-1β, IL-6 and TNF-α) production, and nuclear translocation of p65 and c-JUN. | [75] | 2+ chelating activity was exhibited by peptides. | [67] | |||
| Rice bran | - | KHNRGDEF | Human umbilical vein endothelial cells (HUVECs) | H2O2-induced HUVECs oxidant injury was protected by rice bran peptide (0.1 mM) supplementation via TLR4 binding, pathway inhibition, and suppression of NF-κB activation. | Rice | Alcalase-assisted electron beam irradiation (EBI) | Hydrolysate | EBI treatment at 50 kGy improved DPPH (96.81%) and ABTS (92.04%) radical scavenging activity. | [63] | |
| Corn gluten meal | Ultrasonication assisted alcalase hydrolysis | SGV, LPF, LLPH, LLPF, FLPF, AHL, LGV (<1 kDa) | Ultrasonic pretreatment (5 W/L, 2 s/2 s on/off, 50 °C, and 25 min) significantly increased DH, DPPH and hydroxyl radical scavenging activity and enhanced formation of small size peptides. | [68] |
3. Potential Mechanisms of Cereal Peptidic Antioxidants in MetS Prevention
Recent increase in MetS worldwide and its negative impact on quality of life has led to the search for food ingredients or materials with strong oxidative stress prevention properties. Owing to this, cereal antioxidant peptides have gained great attention among researchers. The amino acid composition, sequence and molecular weight are key factors that contribute to the antioxidant activities of peptides [69][80]. Due to the proteolytic actions of GI enzymes (pepsin, trypsin and chymotrypsin), BP may retain or lose their specific activities at targets sites upon oral ingestion. As such, in vitro activities may not necessarily translate into in vivo efficacies [53][155]. Over the years, the potential antioxidant mechanisms of peptides have mainly been investigated using in vitro chemical assays, in vitro cellular assays, and in vivo animal studies. Although in vivo mechanisms are not fully understood, the activation of the endogenous antioxidant defense system have been reported in vivo [31][135]. Radical scavenging properties, chelation of metal ions, and inhibition of lipid peroxidation assays are the in vitro chemical evaluation methods used to assess antioxidant capacities of peptides and other compounds. The two main mechanisms by which free radicals are deactivated in vitro are the hydrogen atom transfer (HAT) and single electron transfer (SET) [70][171]. The SET mechanism methods frequently used are the DPPH radical scavenging ability and the Trolox equivalent antioxidant capacity (TEAC), while oxygen radical absorption capacity (ORAC), total free radical capture antioxidant parameters (TRAP), and carotene bleach analysis utilizes the HAT mechanism. Ferric reducing antioxidant power (FRAP) and thiobarbituric acid (TBARS) are the most commonly used methods for evaluating metal ion chelation and lipid peroxidation inhibition of antioxidants, respectively [31][135]. Cell-based antioxidant evaluation assays provide a direct reflection of the cytoprotective abilities exerted on oxidation-induced damaged cells, compared to in vitro chemical methods (Table 36). Different human and non-human cell lines are commonly used to assess the antioxidant properties of peptides. Wang et al. [71][172] evaluated the underlying antioxidative mechanism of ADWGGPLPH, a wheat germ-derived peptide on hyperglycemia-induced oxidative stress in vascular smooth muscle cells (VSMCs). ADWGGPLPH significantly prevented cell proliferation induced by high glucose, reduced intracellular ROS production, and suppressed NOX4 protein expression (a key enzyme related to ROS production in vascular cells) via the PKCζ/AMPK signaling pathway. Furthermore, ADWGGPLPH treatment at 4 mg/kg, administered intraperitoneally, enhanced antioxidant abilities by reducing liver MDA levels and increasing SOD expression and attenuated inflammatory cytokine (plasma levels of TNF-α and IL-1β) generation in STZ-induced C57BL/6 diabetic mice. Thus, ADWGGPLPH as a dietary antioxidant supplement could be used to complement antihyperglycemic drugs in the treatment of diabetic vascular complications.
Table 3. Cellular antioxidant effects of cereal peptides and hydrolysates.
| Protein Source | Enzyme | Peptide Sequence/Hydrolysate | Cellular Model | Cellular Outcome | Reference |
|---|---|---|---|---|---|
| Rice | |||||
| [ | |||||
| 76 | |||||
| ] | |||||
| Rice | - | OP60 commercial peptide | HepG2 cells | H2O2- or APAP-induced HepG2 cytotoxicity was reduced by 5 mg/mL OP60 pretreatment via glutathione homeostasis restoration and increased mRNA expression of antioxidant enzymes. | [77] |
| Corn | Alcalase | Zein hydrolysate/peptides | HepG2 cells | Hydrolysate showed higher ORAC activity than native proteins. Peptides (1155.56–1781.63 ng/mL IC50) induced apoptosis at 24 hr by increasing caspase 3 expression. | [78] |
| Corn germ meal | Alcalase | MGGN, MNN, MEN | HepG2 cells | Peptides (0.2 mM) significantly reduced ROS generation in H2O2-induced HepG2 cells. MNN showed the highest cellular antioxidant activity. | [79] |
| Corn gluten meal | Alcalase | corn gluten hydrolysate (CGH1) <1 kDa | HepG2 cells | CGH1-pretreated cells at 2.5 mg/mL upregulated the genes GPX3, GPX5, SOD3, CYGB, SEPP1, and MT3 involved in antioxidant defense. CGH1 suppressed EPHX2 expression, increased cellular EETs, EET-phospholipids formation, and, thus, protected against H2O2-induced HepG2 cell damage. | [80] |
| Corn gluten meal | Alcalase | GLLLPH | HepG2 cells | Corn peptide fractions (CPF) at 2.50 mg/mL exhibited high cellular antioxidant activities and increased the levels of intracellular antioxidant enzymes (SOD, CAT, GR and GSH) in oxidized HepG2 cells. | [81] |
Although numerothough numerous studies have demonstrated the direct antioxidant actions of peptides in vitro using chemical assays, there is still limited understanding of their indirect activation of antioxidant and detoxifying molecular pathways in vivo (Table 47). Presently, the Keap1-Nrf2 signaling pathway is considered one of the plausible antioxidant mechanisms of peptides in vivo. Nuclear factor erythroid 2-related factor 2 (Nrf2) regulates cellular responses against environmental stresses, and is bound to Kelch-like ECH associated protein 1 (Keap1) in the cytoplasm under basal conditions. However, during oxidative stress conditions, Nrf2 is released from Keap1 and translocated into the nucleus, where it binds to antioxidant response elements (AREs) and upregulates target genes [82][83][173,174]. The indirect antioxidative effects of peptides have been shown to be mediated through the Keap1-Nrf2 pathway. The activation of Nrf2 protects cells from oxidative damage via the upregulation of antioxidant and detoxifying enzymes, such as heme oxygenase-1 (HO-1), glutathione reductase (GR), and NAD(P)H quinone oxidoreductase1 (NQO1). Oryza Peptide-P60 (OP60), a commercial rice peptide, was reported to increase intracellular glutathione levels. Moritani et al. [77][175] evaluated the mechanisms underlying the antioxidant potential of this peptide in HepG2 cells and mice models. Their results revealed the cytoprotective effect of OP60 via the Nrf2 signaling pathway. OP60 pretreatment of HepG2 cells at 5 mg/mL reduced cytotoxicity caused by H2O2 or acetaminophen (APAP). The mRNA level of genes encoding heavy and light subunits of γ-glutamylcysteine synthetase (γ-GCS) and other antioxidant enzymes were increased by OP60 treatment, as well as the promotion of Nrf2 nuclear translocation. OP60 treatment at 500 mg/kg in mice prevented oxidative stress-induced liver injury by increasing glutathione levels, heavy subunits of γ-GCS, and heme oxygenase-1 expression in the liver. Thus, OP60 could be utilized as a functional food or ingredient for the prevention or management of MetS associated with oxidative stress. Findings from Sullivan et al. [75][176] revealed an association between the anti-inflammatory potential of sorghum kafirin in LPS-induced inflammation in THP-1 cells, and a reduction of intracellular ROS production. This association may be due to the possibility of kafirin’s ability to directly bind to LPS and inhibit LPS binding to its receptor, TLR4, in kafirin-treated THP-1 cells, thus reducing ROS production induced by LPS. Moreover, reduced secretion of pro-inflammatory cytokines (IL-1β, IL-6 and TNF-α) could be due to decreased nuclear translocation of p65 NF-κB subunits and c-JUN, as a result of limited phosphorylation of ERK 1/2 and JNK caused by a reduction in intracellular ROS production.
Table 4. Antioxidant effects of cereal-derived peptides and hydrolysates in animal models.
| Protein Source | Peptide/Hydrolysate | Purpose of Study | In Vivo Outcome | BSA-Based Human Equivalent Dose (mg/kg) | Reference |
|---|---|---|---|---|---|
| Wheat bran | NL, QL, FL, HAL, AAVL, AKTVF, and TPLTR | Investigated the blood pressure lowering effects of wheat protein hydrolysates and peptides in spontaneous hypertensive rats (SHR). | Oral administration of <1 kDa peptides at 100 mg/kg showed a better reduction of systolic blood pressure (−35 mmHg) after 6 h compared to hydrolysate (−20 mmHg). | 16.22 | [84] |
| Wheat bran | LRP, LQP | Evaluated the effects of peptides on oxidative stress and the AMPK signaling pathway in HFD-induced NASH C57BL/6 mice. | NASH mice treated with 0.20% LRP showed a remarkable decrease in serum d-ROM and a significant increase in BAP levels. Administration of 0.20% LQP increased phospho-AMPK expression and decreased phospho-ACC expression, thus alleviating the severity of NASH. | 7.46 | [85] |
| Wheat bran | ADWGGPLPH | Assessed the antioxidative and antidiabetic vascular dysfunction effects of peptide in STZ-induced C57BL/6 mice. | Administered peptide at 4mg/kg enhanced SOD expression and total antioxidant capacity, and also attenuated hyperglycemia-induced inflammatory factors, such as TNF-α and IL-1β. | 0.32 | [71] |
| OP60 commercial peptide | Evaluated the protective effect of OP60 against APAP-induced hepatic injury in mice. | GSH synthesis and antioxidant enzymes were induced by OP60 (500 mg/kg) via activation of the Nrf2 pathway. | 40.54 | [77] | |
| Corn Gluten meal (CGM) | CGM peptides (<10 kDa) | Investigated the antioxidant capacity of CGMP produced by solid–state fermentation with Bacillus subtilis MTCC5480 in aging rats induced with D-galactose. | Serum and liver antioxidant enzymes (SOD, catalase, glutathione peroxidase) and total antioxidant capacity activities increased, with a decrease in MDA in D-galactose-induced aging rats fed CGMP (250 mg/kg bw). | 40.54 | [86] |
| Corn germ meal | Albumin peptides fractions (APF-4) | Examined the hepatoprotective effects of APF-4 in alcohol-induced liver injury in mice. | APF-4 (800 mg/kg bw) administration significantly reduced activities and levels of hepatic (AST), ALT and MDA, and increased activities of SOD, CAT and GSH. | 64.86 | [87] |
The inhibitie inhibition on of Toll-like receptor 4 (TLR4) pathways and the suppression of NF-κB activation is one of the potential mechanisms by which antioxidative peptides exert their protective functions. The antioxidant effect of rice-derived bran bioactive peptides (RBAP), KHNRGDEF, on H2O2-induced oxidative injury in human umbilical vein endothelial cells (HUVECs) was evaluated by Liang et al. [76][177]. RBAP protected HUVECs against H2O2-induced oxidant injury by binding to, and inhibiting, Toll-like receptor 4 (TLR4) pathways and suppressing NF-κB activation. Cruz-Chamorro et al. [73][178] reported on the reduction of Type 1 T helper (Th1) and Th17 pro-inflammatory cytokines production, suggesting an improvement in cellular anti-inflammatory microenvironment by increasing Th2/Th1 and Th2/Th17 balances in phytohaemagglutinin-P (PHA)-stimulated human peripheral blood mononuclear cells (PBMCs) using alcalase-generated wheat germ protein hydrolysates (WGPHs) treatment. WGPHs improved the total antioxidant capacity by directly scavenging free radicals, increasing the reduced form of glutathione (GSH) levels, and reducing nitric oxide (NO) overproduction. The antioxidative and anti-inflammatory effects of three peptides (MPH-A-I, IALLIPF, and PFLF) from foxtail millet, obtained by alcalase hydrolysis, was evaluated in H2O2-induced human keratinocyte HaCaT cells and RAW264.7 murine macrophages [74][179]. Peptides (IALLIPF and PFLF) from millet prolamins demonstrated promising antioxidant activity by effectively reducing ROS production in H2O2-induced HaCaT cells, inhibiting MDA production, and increasing GSH levels more than the peptide fraction of MPH-A-I. Furthermore, peptides suppressed the production of NO and pro-inflammatory cytokines (TNF-α, IL-1β and IL-6) in (lipopolysaccharide) LPS-stimulated RAW264.7 cells. In addition, pretreatment with millet peptides was found to suppress the production of phosphorylated proteins (P-p38, P-Erk 1/2, and P-JNK) in the MAPK pathway. The potential antioxidant and anticancer activities of papain–hydrolyzed sorghum kafirin peptide antioxidants in human hepatocarcinoma (HepG2) cells have been demonstrated by Xu et al. [89][132]. They found that after 72 h treatment, antioxidant peptides (1–3 kDa) at 50 and 200 μg/mL significantly inhibited the growth of HepG2 cells, without any negative effect on cell viability. Findings from Wang et al. [80][180] showed intracellular ROS scavenging activities and the regulation of antioxidant defense and ROS metabolism relevant gene expressions in HepG2 cells by alcalase-derived corn gluten hydrolysate (CGH). Pretreatment of cells with CGH1 enhanced the expression of several genes (GPX3, GPX5, SOD3, CYGB, SEPP1, and MT3) initially suppressed in H2O2-induced HepG2 oxidative damaged cells. The suppression of EPHX2 expression by CGH1, which could be attributed to its antioxidant effect, was associated with arachidonic acid metabolism shown by an increase in cellular epoxyeicosatrienoic acid (EET) and EET-phospholipids formation. Miscalculation and misinterpretation of effective animal doses are two of the main problems encountered in interspecies comparisons and safe starting dose determination for initial clinical trials. Reagan-Shaw et al. [88][181] proposed the use of a body surface area (BSA) normalization method as an appropriate means for translating doses from animal studies to human equivalent doses (HEDs); instead of using a simple conversion based on body weight, which oftentimes results in misinterpretation and inappropriate comparisons between species due to invalid or inaccurate calculations. The authors suggested BSA as a suitable conversion factor for clinical trials, as it correlates well with several mammalian biological parameters, such as oxygen utilization, basal metabolism, caloric expenditure, blood volume, circulating plasma proteins, and renal function. However, for an average 60 kg person, an estimated dose of 3891.6 mg, 2432.4 mg, and 972 mg is required for corn germ albumin peptide fraction 4, OP60 rice commercial peptide, corn gluten meal peptides, and wheat bran peptides HEDs of 64.86, 40.54, 16.22 mg/kg, respectively (Table 47). These concentrations are not reasonably obtainable compared to a dose of 19.2 mg and 447.60 mg for wheat bran peptides, ADWGGPLPH and LRP, with an HED of 0.32 and 7.46 mg/kg, respectively. In addition, the size of the peptide, the mode of administration—whether oral, injection or perfusion—and the delivery system have a great impact on peptide stability, transport, and bioavailability.
