You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Giuditta Dal Cortivo.
Calcium- and integrin-binding protein 2 (CIB2) is a small EF-hand protein capable of binding Mg2+ and Ca2+ ions. Its biological function remains largely unclear, an increasing number of studies have shown that CIB2 is an essential component of the mechano-transduction machinery that operates in cochlear hair cells. In addition, CIB2 has been implicated in a multitude of very different processes, ranging from integrin signaling in platelets and skeletal muscle to autophagy, suggesting extensive functional plasticity.
- non-syndromic deafness
- Usher syndrome
- hearing
- integrin signaling
1. Introduction
More than two decades ago, Seki et al. isolated from a cDNA library of human fetal brains a clone encoding a 187 amino acid protein, initially named kinase interacting protein 2 and currently referred to as calcium-and integrin-binding protein 2 (CIB2)[1]. Examination of the transcript highlighted a broad distribution in various human tissues, including brain (both fetal and adult), heart, kidney, lung, thymus, spleen, placenta, ovary, and testis, thus suggesting that the product of the CIB2 gene is involved in basic cellular functions[1]. Further studies also confirmed a broad expression of CIB2 in mice, specifically in the inner ear, outer and inner retina, retinal pigmented epithelium[2], and skeletal muscle, with higher expression levels in sarcolemma, the myotendinous junction, and the neuromuscular junction in adult mice and lower expression in brain and lungs[3]. Evidence of CIB2 expression has also been reported in adult rat brain, with specific localization in the hippocampus and in the sensory, entorhinal, and prefrontal cortex, with significant intracellular localization at the Golgi apparatus and neurites[4]. High expression levels of the ortholog CIB2 in sheep have been found by RT-PCR in different tissues, mainly stomach, heart, and ovary[5].
The extremely wide expression of CIB2 in various tissues of different organisms suggests its implication in a wide variety of biochemical processes. A broad and multifaceted biological function is consistent with its membership in the CIB family of Ca2+- and Mg2+-binding proteins[6]. This family includes homologous proteins showing evolutionary relationships with the class of neuronal calcium sensors (NCS)[7]. The biological role of CIB2 was initially associated with its interaction with specific integrins involved in intra- and extracellular signaling pathways[3][8]. More recently, however, increasing lines of evidence have associated CIB2 with hearing physiology and pathology[9], suggesting that CIB2 is an essential component for the normal development of hair cells in the inner ear and possibly a structural component of the hair cell mechanotransduction complex[10][11]. In parallel, biochemical and biophysical studies[12][13][14] have highlighted similarities but also important differences between CIB2 and other members of the CIB family, which should be considered when investigating its versatile and still largely unknown functions and involvement in processes as diverse as autophagy[15], cancer[16], and muscular dystrophy[3].
2. Structural, Biochemical, and Biophysical Properties of CIB2
The discovery of the CIB gene family dates back to the 1990s, with the identification of the first member named calcium- and integrin-binding protein (CIB1)[17] or KIP (kinase interacting protein)[18] based on the putative function, which showed 58% sequence similarity with calcineurin B and 56% with calmodulin. Since then, several structural studies[13][19][20] have clarified the features shared by the CIB family members, which contain three further homologs, CIB2, CIB3, and CIB4, in the human genome, and highlighted commonalities with the neuronal calcium sensor proteins (NCS) family. Human CIB2 (NG_033006) is a 33,929 bp gene located on chromosome 15 (15q25.1) encoding four different isoforms, consisting of 4–6 exons [2]. The canonical sequence (Uniprot code: O75838-1) encodes a 187 amino acid protein of 21.6 kDa harboring two functional EF-hands, the metal-binding helix–loop–helix motif characterizing the largest family of Ca2+-binding proteins in eukaryotic cells[21]. Functional EF-hands are located at the C-terminal domain of CIB2 and will be referred to as EF3 and EF4 in this revisewarch to distinguish them from the non-functional EF1 and EF2 located in the N-terminal domain (Figure 1). The three-dimensional structure of CIB2 is currently unknown, although homology models have been built based on the experimental structure of CIB1[14] and CIB3[20].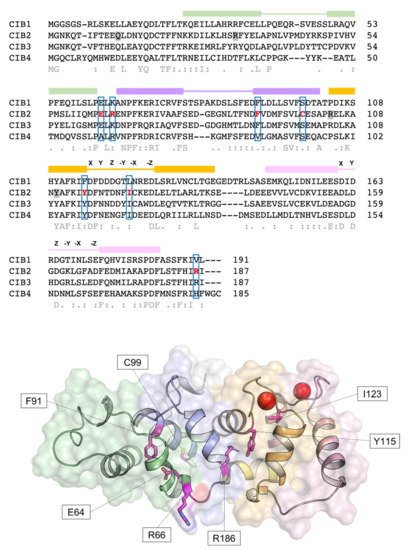
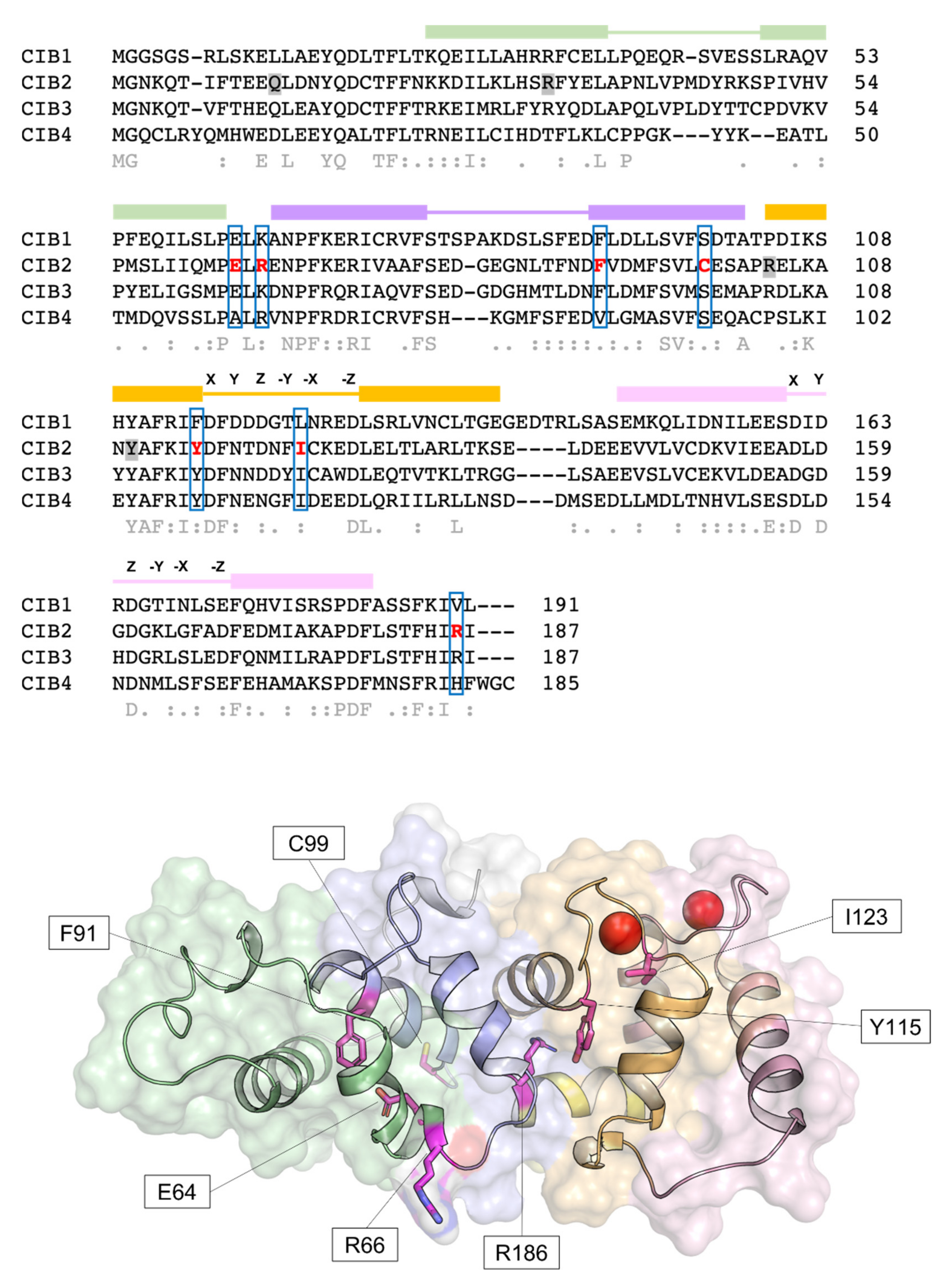 Figure 1. Sequence and structural properties of CIB2 and its pathogenetic variants. Top: multiple amino acid sequence alignment of human CIB1, CIB2, CIB3, and CIB4 performed by T–Coffee (https://tcoffee.crg.eu/apps/tcoffee/index.html, accessed on 22 February 2022). Consensus sequence and similarity descriptors are reported in grey text. Residues target of pathogenetic point mutations are grey-shaded in case of nonsense mutations or marked in red and blue-boxed in case of missense mutations. EF-hand motifs are displayed by colored boxes, and residues involved in Ca2+-coordination are indicated by letters referring to the canonical pentagonal bipyramidal geometry on the respective loops. Bottom: Representation of the three-dimensional structure of human CIB2 based on the homology model used in [14]. Cartoon and surface representation are superposed. The N-terminal region is colored gray, while the C-terminal region (helix 10) is colored yellow. EF1, EF2, EF3, and EF4 are colored green, slate blue, orange, and pink, respectively. Residue targets of pathogenic missense mutations are represented by magenta sticks, and Ca2+ ions are represented by red spheres.
Figure 1. Sequence and structural properties of CIB2 and its pathogenetic variants. Top: multiple amino acid sequence alignment of human CIB1, CIB2, CIB3, and CIB4 performed by T–Coffee (https://tcoffee.crg.eu/apps/tcoffee/index.html, accessed on 22 February 2022). Consensus sequence and similarity descriptors are reported in grey text. Residues target of pathogenetic point mutations are grey-shaded in case of nonsense mutations or marked in red and blue-boxed in case of missense mutations. EF-hand motifs are displayed by colored boxes, and residues involved in Ca2+-coordination are indicated by letters referring to the canonical pentagonal bipyramidal geometry on the respective loops. Bottom: Representation of the three-dimensional structure of human CIB2 based on the homology model used in [14]. Cartoon and surface representation are superposed. The N-terminal region is colored gray, while the C-terminal region (helix 10) is colored yellow. EF1, EF2, EF3, and EF4 are colored green, slate blue, orange, and pink, respectively. Residue targets of pathogenic missense mutations are represented by magenta sticks, and Ca2+ ions are represented by red spheres.
2.1. CIB2 Has the Potential to Work as a Ca
2+
and Mg
2+
Sensor Protein
Since its discovery and based on the homology with CIB1, CIB2 was expected to act as a Ca2+ sensor, thereby changing its conformation upon the binding of Ca2+ ions and possibly other cations and acquiring a structure that allows the regulation of specific molecular targets. Human CIB2 was, therefore, heterologously expressed in E. coli and purified to study its structural and biophysical properties. Huang et al. confirmed that CIB2 acts as a typical EF-hand calcium sensor, which undergoes a significant Ca2+ induced conformational change, thereby exposing a hydrophobic patch and increasing the content of secondary structure—essentially α-helix—as proven by far-UV circular dichroism (CD) spectroscopy[13]. Interestingly, the protein was also shown to respond to Mg2+ with similar intensity, although 8-anilinonaphthalene-1-sulfonic acid (ANS) fluorescence showed that the most prominent conformational change in terms of exposure of hydrophobic surface was observed in the presence of Ca2+, a result confirmed by a subsequent independent study[14]. Nuclear magnetic resonance (NMR) spectroscopy (1H-15N-HSQC NMR) showed that in the absence of any cation (apo- condition), the structure of CIB2 is not ordered, while far-UV CD spectroscopy demonstrated residual α-helix secondary structure. The addition of either Mg2+ or Ca2+ induced a well-folded tertiary structure in CIB2, with many peaks overlapping despite the detectable structural difference among the cation-bound specific state[13]. An independent study was performed under similar conditions, after removing the poly-His-tag used in [13] for protein purification purposes, and led to very similar results, indicating that apo-CIB2 lacks tertiary structure and forms a molten globule state, as also confirmed by near-UV CD spectroscopy[14].2.2. An Alleged Ca
2+
Sensor? CIB2 Constitutively Binds Mg
2+
but Has Low Affinity for Ca
2+
In vitro studies showed that all CIB proteins respond to Mg2+ and Ca2+ stimuli in a relatively similar manner in terms of general structural features[13]; however, a recent investigation by Vallone et al. highlighted important peculiarities of CIB2 in cation-sensing[14]. NMR titration experiments reporting on the Ca2+ binding state of specific residues suggested that when starting from the apo-protein, EF3 is the first EF-hand in CIB2 to be occupied by Ca2+, followed by EF4. The variation of 1H-15N HSQC peak intensity for the EF4 residue Gly 162 upon Ca2+ titration experiments displays a sigmoidal shape, indicative of positive cooperativity[14]. However, in the presence of physiological levels of free Mg2+ (approximately 1 mM), EF3 seems never to be occupied by Ca2+ but rather constitutively Mg2+-bound. A Mg2+/Ca2+ exchange would thus possibly occur only in EF4[14] (Figure 2).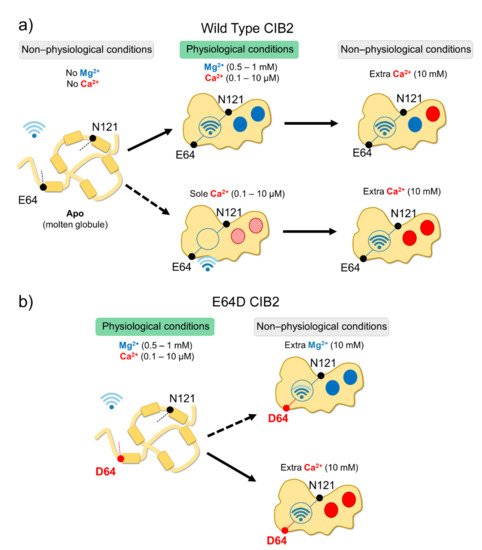
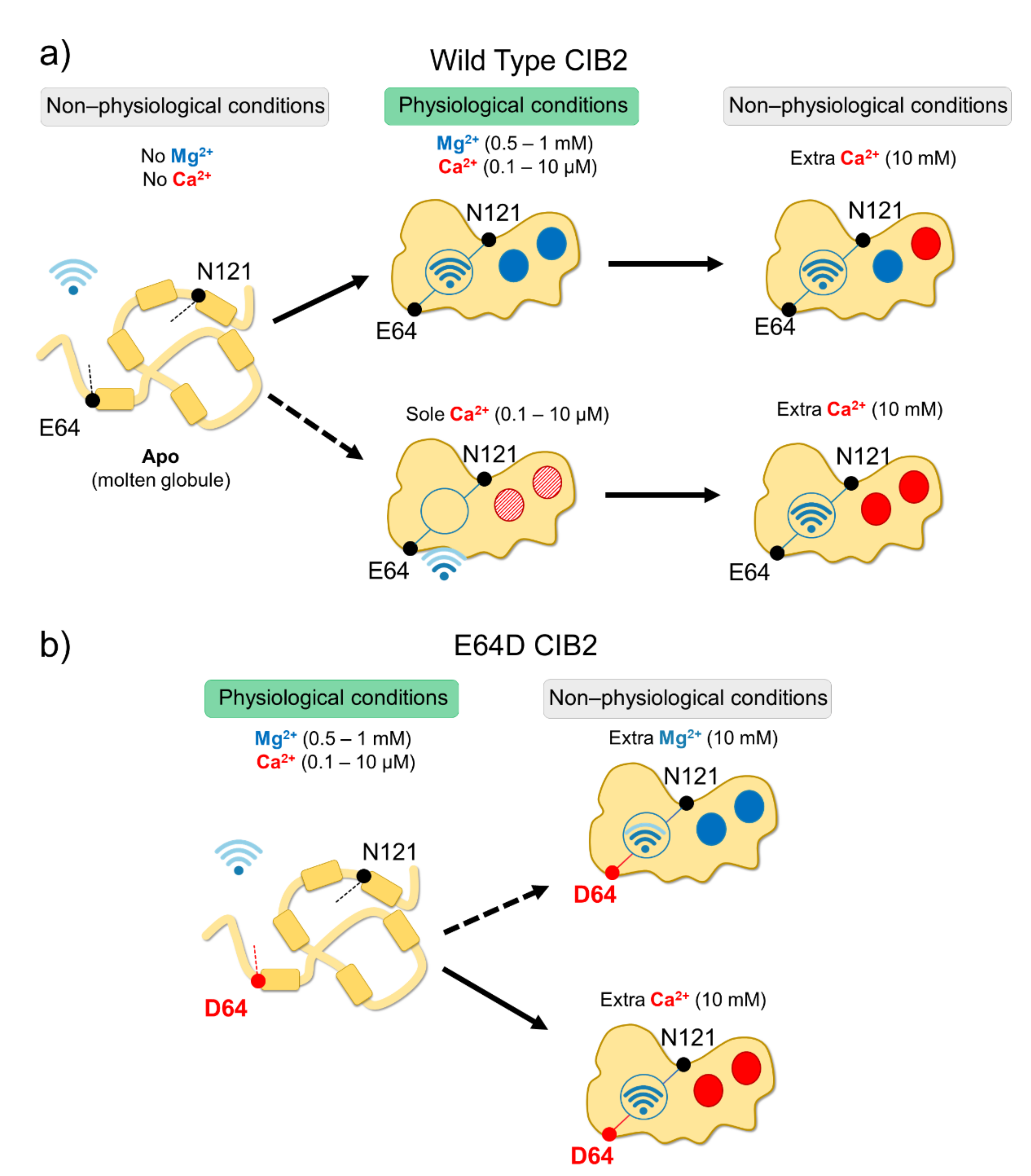 Figure 2. Schematic representation of CIB2 cation sensitivity and structural transitions. (a) In the absence of cations, WT CIB2 is in a molten globule state (left). The addition of a physiological concentration of both Mg2+ and Ca2+ (middle top panel) triggers the binding of Mg2+ in EF3 and EF4. In the presence of physiological Ca2+ and absence of Mg2+ (middle bottom panel), Ca2+ binding is less favorable. When extra (non-physiological) Ca2+ concentrations are added, Ca2+ replaces Mg2+ in EF4 (upper right panel). (b) E64D CIB2 is in a molten globule state under physiological conditions (left). A well-defined three-dimensional structure is acquired upon the addition of extra (non-physiological) Mg2+ (upper right panel) or Ca2+ (bottom right panel), with a more pronounced effect for Ca2+. The Wi-Fi logo represents the strength of allosteric interactions between E64 and N121 residues.
Figure 2. Schematic representation of CIB2 cation sensitivity and structural transitions. (a) In the absence of cations, WT CIB2 is in a molten globule state (left). The addition of a physiological concentration of both Mg2+ and Ca2+ (middle top panel) triggers the binding of Mg2+ in EF3 and EF4. In the presence of physiological Ca2+ and absence of Mg2+ (middle bottom panel), Ca2+ binding is less favorable. When extra (non-physiological) Ca2+ concentrations are added, Ca2+ replaces Mg2+ in EF4 (upper right panel). (b) E64D CIB2 is in a molten globule state under physiological conditions (left). A well-defined three-dimensional structure is acquired upon the addition of extra (non-physiological) Mg2+ (upper right panel) or Ca2+ (bottom right panel), with a more pronounced effect for Ca2+. The Wi-Fi logo represents the strength of allosteric interactions between E64 and N121 residues.
2.3. An Inter-Domain Allosteric Switch Regulates the Conformational Transitions of CIB2
Although with a lower affinity than CIB1, CIB2 binds Mg2+ and Ca2+ (the latter under non-physiological conditions), and this significantly stabilizes the structure of the protein. Thermal denaturation studies performed by monitoring the protein secondary structure content by far-UV CD spectroscopy indeed proved that apo-CIB2 is rather unstable, with a melting temperature (Tm) of 35 °C, while the addition of Mg2+ or Ca2+ significantly increases the stability by enhancing the Tm of 11 and 8 °C, respectively[14]. In the co-presence of both cations, the Tm resembles that of Mg2+, confirming the minor stabilizing role of Ca2+ compared to Mg2+. The stabilizing effects of cations on the protein tertiary structure are even more apparent when analyzed by NMR, which also highlighted an important allosteric mechanism connecting EF3 with the non-functional EF1 motif. An inter-domain allosteric communication was detected between the EF3 binding loop and the residue E64, which is predicted to form an electrostatic interaction with R33, thereby contributing to the stability of the EF1 subdomain. This was clearly demonstrated by 1H-15N HSQC NMR spectra, where the chemical shift of N121 in EF3 and E64 in EF1 were found to show the same pattern upon Ca2+-titration experiments. The switch that allows CIB2 to acquire a functional conformation at physiological levels of Mg2+ seems, therefore, to be finely regulated by an allosteric, long-range communication connecting EF1 with EF3 (Figure 2a). Interestingly, E64 is mutated into an Asp (E64D) in a group of patients affected by Usher syndrome type 1 J[2]. Vallone et al. demonstrated that the apparently conservative E64D substitution breaks up such inter-domain communication, resulting in a protein that is unable to bind Mg2+, which is necessary to adopt the required physiological conformation, thus providing a first mechanistic explanation for the molecular basis of disease[14].2.4. CIB2 Is Monomeric, but It Could Dimerize in the Presence of a Target
Analytical size exclusion chromatography (SEC) is a valuable technique to study the hydrodynamic properties of proteins, and it can be used to assess the molecular weight (MW) of the eluting protein when a calibration curve is obtained in the same conditions for a number of globular proteins is available[34]. The apparent MW of CIB2 obtained by SEC in the presence of Mg2+ or co-presence of Mg2+ and Ca2+ was determined to be around 39 kDa, therefore significantly higher the theoretical MW of a monomer (22 kDa); this was initially interpreted as evidence of the dimeric nature of CIB2[14]. A similar conclusion was supported by dynamic light scattering (DLS) measurements. A subsequent study employing a variety of mass spectrometry (MS)-based techniques, including native ESI-MS, MALDI-TOF-MS, and cross-linking/MS integrated with novel SEC and DLS experiments based on a more accurate selection of the heterogeneous components of the elution bands, demonstrated that CIB2 was monomeric under all tested conditions[12]. A comparison with results obtained with recoverin and calmodulin suggested that the apparent MW, extrapolated by analytical SEC for a Ca2+ sensor protein and based on the hydrodynamic radius, can significantly differ from the real value when the protein has a high hydrophobic solvent-accessible surface, such as in the case of recoverin and CIB2[12]. Analytical SEC could then be driven to erroneous conclusions as to the oligomeric state of the protein[14]. This can explain the apparent contradiction with previous studies that detected CIB2 dimers based on FRET and co-immunoprecipitation. In these studies, CIB2 was fused to GFP[2][35] or tdTomato fluorescent protein[2]; its uncommon hydrodynamic properties, together with a possible interference of the bulky fusion constructs, may have led to the erroneous detection of a dimer[12]. The most recent and thorough dedicated investigation performed with untagged purified CIB2 seems to exclude the existence of CIB2 dimers under conditions mimicking the physiological ones[12]. In this respect, the oligomeric state of CIB2 resembles that of CIB1. Ca2+-bound CIB1 was indeed found to be monomeric in the crystallographic structure reported by Gentry et al.[19], and this finding has been supported by NMR diffusion, SEC, and sedimentation equilibrium experiments[36][37]. The head-to-tail dimer reported by another crystallographic study could result from the specific conditions for crystal formation, which included a GSH moiety at the N-domain[38]. Surface plasmon resonance (SPR) spectroscopy also excluded the dimerization of CIB2 over a broad range of conditions, including selective incubation with Mg2+ and Ca2+[12], but suggested an interesting mechanism of binding to integrin. The formation of a protein–peptide complex between CIB2 and a peptide from the α7B integrin was shown to possibly drive the binding of a second CIB2 molecule; the process seems to be kinetically favored in the sole presence of Mg2+[12]. Although this hypothesis awaits confirmation, the mechanism of target-induced CIB2 dimerization would explain the 2:1 protein:peptide stoichiometry detected for the same interaction in a previous fluorescence study [14], and it is tempting to speculate that it might play a role in the integrin signaling mediated by CIB proteins.2.5. CIB2 Myristoylation
Myristoylation is a post-translational modification operated by the N-myristoyl transferase that, in vivo, covalently binds a 14-carbon saturated fatty acid (myristoyl moiety) to the N-terminal glycine of proteins harboring the consensus sequence MGXXXS/T[39]. CIB1, CIB2, and CIB3 have the optimal consensus sequence (Figure 1) and are likely to be myristoylated in vivo, while CIB4 is not. The binding of Ca2+ ions to some NCS proteins triggers the so-called “myristoyl switch” mechanism, which extrudes the myristoyl moiety from a hydrophobic cleft in the protein milieu to a fully solvent-exposed state, thus allowing membrane-binding and permitting specific cell localization and target interaction[40]. It is not clear whether CIB1 undergoes a Ca2+-induced myristoyl-switch, as contrasting conclusions have been reported by different groups. Jarman et al.[41] concluded that the myristoyl switch mechanism occurs in CIB1, and it is necessary to translocate the sphingosine kinase 1 (SK1) target to the membrane; in contrast, according to Blazejczyk et al., the myristoyl moiety of CIB1 is likely solvent-exposed rather than buried within the protein, regardless of its Ca2+ binding state[42]. Whether or not the Ca2+-induced switching mechanism occurs, the myristoylation of CIB1 is still important for protein stability and membrane targeting[43][44], as well as shuttling its binding partners to the membrane[41][45]. Myristoylation-deficient CIB1 variants retain a high affinity for target proteins and peptides both in vitro and in vivo[42][44][46][47], thus suggesting that the myristoyl group is not specifically involved in target recognition. Much less information is available about the myristoylation of CIB2. Blazejczyk et al.[4] expressed GFP-tagged CIB2 and a myristoylation-blocking variant (G2ACIB2-GFP) in COS-7 cells in the presence of radioactive 3H-myristic acid and compared the effect of CIB2 myristoylation with that of VILIP1-GFP[48], known to undergo the Ca2+-induced myristoyl switch, thereby resulting in specialized membrane compartment localization. In contrast to what was observed for VILIP1-GFP, no Ca2+-dependent translocation was detected for CIB2-GFP, which was interpreted as a lack of Ca2+-induced myristoyl switch[4]. Moreover, localization of CIB2 was essentially limited to the crude membrane fraction, regardless of the tag (CIB2-FLAG gave the same results) and, surprisingly, of myristoylation as the same behavior was detected for G2ACIB2-GFP[4]. This would exclude a direct association of CIB2 to the membrane by myristoyl anchoring and rather suggests that the binding is mediated directly by lipids or by an interaction with other proteins. Within COS-7 cells, CIB2 co-localizes with the Golgi apparatus and not with the nucleus, and the absence of myristoylation does not affect this pattern[4]. An independent study by Zhu et al.[49] confirmed that CIB2 does not undergo a Ca2+-induced myristoyl switch and blocks the agonist-induced membrane translocation of SK1, at odds with the activity of CIB1. Experiments were performed with HA-tagged CIB2 in HEK293 cells labeled with 3H-myristic acid and showed that the interaction of CIB2 with SK1 was independent of the presence of Ca2+ or Mg2+. Further experiments are needed to elucidate the biological role of CIB2 myristoylation, and biophysical studies may clarify the structural and mechanistic aspects that remain somewhat unclear.References
- Naohiko Seki; Astushi Hattori; Akiko Hayashi; Sumie Kozuma; Miki Ohira; Tada-Aki Hori; Toshiyuki Saito; Structure, expression profile and chromosomal location of an isolog of DNA-PKcs interacting protein (KIP) gene. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1999, 1444, 143-147, 10.1016/s0167-4781(98)00253-x.
- Saima Riazuddin; Inna A Belyantseva; Arnaud P J Giese; Kwanghyuk Lee; Artur A Indzhykulian; Sri Pratima Nandamuri; Rizwan Yousaf; Ghanshyam P Sinha; Sue Lee; David Terrell; et al.Rashmi S HegdeRana A AliSaima AnwarPaula B Andrade-ElizondoAsli SirmaciLeslie V PariseSulman BasitAbdul WaliMuhammad AyubMuhammad AnsarWasim AhmadShaheen N KhanJaved AkramMustafa TekinSheikh RiazuddinTiffany CookElke K BuschbeckGregory I FrolenkovSuzanne M LealThomas B FriedmanZubair M Ahmed Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nature Genetics 2012, 44, 1265-1271, 10.1038/ng.2426.
- Mattias Häger; Maria Giulia Bigotti; Renata Meszaros; Virginie Carmignac; Johan Holmberg; Valérie Allamand; Mikael Åkerlund; Sebastian Kalamajski; Andrea Brancaccio; Ulrike Mayer; et al.Madeleine Durbeej Cib2 Binds Integrin α7Bβ1D and Is Reduced in Laminin α2 Chain-deficient Muscular Dystrophy. Journal of Biological Chemistry 2008, 283, 24760-24769, 10.1074/jbc.m801166200.
- Magdalena Blazejczyk; Adam Sobczak; Katarzyna Debowska; Marta Wisniewska; Aneta Kirilenko; Sławomir Pikuła; Jacek Jaworski; Jacek Kuznicki; Urszula Wojda; Biochemical characterization and expression analysis of a novel EF-hand Ca2+ binding protein calmyrin2 (Cib2) in brain indicates its function in NMDA receptor mediated Ca2+ signaling. Archives of Biochemistry and Biophysics 2009, 487, 66-78, 10.1016/j.abb.2009.05.002.
- Yan Yu; Xuemei Song; Lixin Du; Chuduan Wang; Molecular characterization of the sheep CIB1 gene. Molecular Biology Reports 2008, 36, 1799-1809, 10.1007/s11033-008-9383-4.
- AP Yamniuk, HJ Vogel - Calcium Binding Proteins, 2006; Review Insights into the Structure and Function of Calcium-and Integrin-Binding Proteins. Calcium Bind. Proteins 2006, 1, 150-155.
- Daniele Dell'orco; Karl-Wilhelm Koch; Michael R. Kreutz; Jose R. Naranjo; Beat Schwaller; Editorial: Neuronal Calcium Sensors in Health and Disease. Frontiers in Molecular Neuroscience 2019, 12, 278, 10.3389/fnmol.2019.00278.
- Jan C. Denofrio; Weiping Yuan; Brenda R. Temple; Holly R. Gentry; Leslie V. Parise; Characterization of calcium- and integrin-binding protein 1 (CIB1) knockout platelets: Potential compensation by CIB family members. Thrombosis and Haemostasis 2008, 100, 847-856, 10.1160/th08-06-0351.
- Agnieszka Jacoszek; Agnieszka Pollak; Rafał Płoski; Monika Ołdak; Advances in genetic hearing loss: CIB2 gene.. European Archives of Oto-Rhino-Laryngology 2016, 274, 1791-1795, 10.1007/s00405-016-4330-9.
- David P. Corey; Nurunisa Akyuz; Jeffrey R. Holt; Function and Dysfunction of TMC Channels in Inner Ear Hair Cells. Cold Spring Harbor Perspectives in Medicine 2018, 9, a033506, 10.1101/cshperspect.a033506.
- Jeffrey R. Holt; Mélanie Tobin; Johannes Elferich; Eric Gouaux; Angela Ballesteros; Zhiqiang Yan; Zubair M. Ahmed; Teresa Nicolson; Putting the Pieces Together: the Hair Cell Transduction Complex. Journal of the Association for Research in Otolaryngology 2021, 22, 601-608, 10.1007/s10162-021-00808-0.
- Giuditta Dal Cortivo; Valerio Marino; Claudio Iacobucci; Rosario Vallone; Christian Arlt; Anne Rehkamp; Andrea Sinz; Daniele Dell’Orco; Oligomeric state, hydrodynamic properties and target recognition of human Calcium and Integrin Binding protein 2 (CIB2). Scientific Reports 2019, 9, 1-11, 10.1038/s41598-019-51573-3.
- Hao Huang; Joel N. Bogstie; Hans J. Vogel; Biophysical and structural studies of the human calcium- and integrin-binding protein family: understanding their functional similarities and differences. Biochemistry and Cell Biology 2012, 90, 646-656, 10.1139/o2012-021.
- Rosario Vallone; Giuditta Dal Cortivo; Mariapina D'Onofrio; Daniele Dell'orco; Preferential Binding of Mg2+ Over Ca2+ to CIB2 Triggers an Allosteric Switch Impaired in Usher Syndrome Type 1J. Frontiers in Molecular Neuroscience 2018, 11, 274, 10.3389/fnmol.2018.00274.
- Saumil Sethna; Patrick A. Scott; Arnaud P. J. Giese; Todd Duncan; Xiaoying Jian; Sheikh Riazuddin; Paul A. Randazzo; T. Michael Redmond; Steven L. Bernstein; Saima Riazuddin; et al.Zubair M. Ahmed CIB2 regulates mTORC1 signaling and is essential for autophagy and visual function. Nature Communications 2021, 12, 1-19, 10.1038/s41467-021-24056-1.
- Xianwang Wang; Yue Yang; Wen-Qi Cai; Yingying Lu; The Relationship of Sphingosine Kinase 1 With Pyroptosis Provides a New Strategy for Tumor Therapy. Frontiers in Immunology 2020, 11, 574990, 10.3389/fimmu.2020.574990.
- Ulhas P. Naik; Pankaj M. Patel; Leslie V. Parise; Identification of a Novel Calcium-binding Protein That Interacts with the Integrin αIIb Cytoplasmic Domain. Journal of Biological Chemistry 1997, 272, 4651-4654, 10.1074/jbc.272.8.4651.
- Xiantuo Wu; Michael R Lieber; Interaction between DNA-dependent protein kinase and a novel protein, KIP. Mutation Research/DNA Repair 1997, 385, 13-20, 10.1016/s0921-8777(97)00035-9.
- Holly R. Gentry; Alex U. Singer; Laurie Betts; Cheng Yang; Joseph D. Ferrara; John Sondek; Leslie V. Parise; Structural and Biochemical Characterization of CIB1 Delineates a New Family of EF-hand-containing Proteins. Journal of Biological Chemistry 2005, 280, 8407-8415, 10.1074/jbc.m411515200.
- Xiaoping Liang; Xufeng Qiu; Gilman Dionne; Christopher L. Cunningham; Michele L. Pucak; Guihong Peng; Ye-Hyun Kim; Amanda Lauer; Lawrence Shapiro; Ulrich Müller; et al. CIB2 and CIB3 are auxiliary subunits of the mechanotransduction channel of hair cells. Neuron 2021, 109, 2131-2149.e15, 10.1016/j.neuron.2021.05.007.
- Hiroshi Kawasaki; Shigenari Nakayama; Robert H Kretsinger; Classification and evolution of EF-hand proteins. BioMetals 1998, 11, 277-295, 10.1023/a:1009282307967.
- Aaron P. Yamniuk; Leonard T. Nguyen; And Tung T. Hoang; Hans J. Vogel; Metal Ion Binding Properties and Conformational States of Calcium- and Integrin-Binding Protein. Biochemistry 2004, 43, 2558-2568, 10.1021/bi035432b.
- ‡ Aaron P. Yamniuk; ‡ Dylan M. Silver; ‡ Kathryn L. Anderson; § And Stephen R. Martin; ‡ Hans J. Vogel; Domain Stability and Metal-Induced Folding of Calcium- and Integrin-Binding Protein 1. Biochemistry 2007, 46, 7088-7098, 10.1021/bi700200z.
- Sara Linse; A. Helmersson; S. Forsén; Calcium binding to calmodulin and its globular domains. Journal of Biological Chemistry 1991, 266, 8050-8054, 10.1016/s0021-9258(18)92938-8.
- Michael J. Berridge; Martin Bootman; Peter Lipp; Calcium - a life and death signal. Nature 1998, 395, 645-648, 10.1038/27094.
- Karl-Wilhelm Koch; Daniele Dell’Orco; A Calcium-Relay Mechanism in Vertebrate Phototransduction. ACS Chemical Neuroscience 2013, 4, 909-917, 10.1021/cn400027z.
- Karl-Wilhelm Koch; Daniele Dell'Orco; Protein and Signaling Networks in Vertebrate Photoreceptor Cells. Frontiers in Molecular Neuroscience 2015, 8, 67, 10.3389/fnmol.2015.00067.
- Andrea Romani; A. Scarpa; Regulation of cell magnesium. Archives of Biochemistry and Biophysics 1992, 298, 1-12, 10.1016/0003-9861(92)90086-c.
- Romani, A.M.; Scarpa, A.; Regulation of cellular magnesium. Frontiers in Bioscience 2000, 5, 720-734.
- Stéphane M. Gagné; And Monica X. Li; Brian D. Sykes; Mechanism of Direct Coupling between Binding and Induced Structural Change in Regulatory Calcium Binding Proteins. Biochemistry 1997, 36, 4386-4392, 10.1021/bi963076+.
- Zenon Grabarek; Insights into modulation of calcium signaling by magnesium in calmodulin, troponin C and related EF-hand proteins. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2011, 1813, 913-921, 10.1016/j.bbamcr.2011.01.017.
- Steven K. Drake; Joseph J. Falke; Kinetic Tuning of the EF-Hand Calcium Binding Motif: The Gateway Residue Independently Adjusts (i) Barrier Height and (ii) Equilibrium. Biochemistry 1996, 35, 1753-1760, 10.1021/bi952335c.
- Valerio Marino; Matteo Riva; Davide Zamboni; Karl-Wilhelm Koch; Daniele Dell'Orco; Bringing the Ca 2+ sensitivity of myristoylated recoverin into the physiological range. Open Biology 2021, 11, 200346, 10.1098/rsob.200346.
- Valentina La Verde; Paola Dominici; Alessandra Astegno; Determination of Hydrodynamic Radius of Proteins by Size Exclusion Chromatography. BIO-PROTOCOL 2017, 7, 1461, 10.21769/bioprotoc.2230.
- Arnaud Giese; Yiquan Tang; Ghanshyam Sinha; Michael Bowl; Adam Goldring; Andrew Parker; Mary Freeman; Steve D. M. Brown; Saima Riazuddin; Robert Fettiplace; et al.William SchaferGregory I. FrolenkovZubair M. Ahmed CIB2 interacts with TMC1 and TMC2 and is essential for mechanotransduction in auditory hair cells. Nature Communications 2017, 8, 1-13, 10.1038/s41467-017-00061-1.
- Aalim M. Weljie; Aaron P. Yamniuk; Hidenori Yoshino; Yoshinobu Izumi; Hans J. Vogel; Protein conformational changes studied by diffusion NMR spectroscopy: Application to helix-loop-helix calcium binding proteins. Protein Science 2003, 12, 228-236, 10.1110/ps.0226203.
- Adam Sobczak; Magdalena Blazejczyk; Grzegorz Piszczek; Gang Zhao; Jacek Kuznicki; Urszula Wojda; Calcium-binding calmyrin forms stable covalent dimers in vitro, but in vivo is found in monomeric form.. Acta Biochimica Polonica 2005, 52, 469-476, 10.18388/abp.2005_3461.
- Chad J. Blamey; Christopher Ceccarelli; Ulhas P. Naik; Brian J. Bahnson; The crystal structure of calcium- and integrin-binding protein 1: Insights into redox regulated functions. Protein Science 2005, 14, 1214-1221, 10.1110/ps.041270805.
- Megan H. Wright; William P. Heal; David J. Mann; Edward W. Tate; Protein myristoylation in health and disease. Journal of Chemical Biology 2009, 3, 19-35, 10.1007/s12154-009-0032-8.
- James B Ames; Toshiyuki Tanaka; Lubert Stryer; Mitsuhiko Ikura; Portrait of a myristoyl switch protein. Current Opinion in Structural Biology 1996, 6, 432-438, 10.1016/s0959-440x(96)80106-0.
- Kate E. Jarman; Paul A.B. Moretti; Julia R. Zebol; Stuart M. Pitson; Translocation of Sphingosine Kinase 1 to the Plasma Membrane Is Mediated by Calcium- and Integrin-binding Protein 1. Journal of Biological Chemistry 2010, 285, 483-492, 10.1074/jbc.m109.068395.
- Magdalena Blazejczyk; Urszula Wojda; Adam Sobczak; Christina Spilker; Hans-Gert Bernstein; Eckart D. Gundelfinger; Michael R. Kreutz; Jacek Kuznicki; Ca2+-independent binding and cellular expression profiles question a significant role of calmyrin in transduction of Ca2+-signals to Alzheimer's disease-related presenilin 2 in forebrain. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2006, 1762, 66-72, 10.1016/j.bbadis.2005.09.006.
- Jan K. Hennigs; Nicole Burhenne; Frauke Stähler; Marcel Winnig; Bettina Walter; Wolfgang Meyerhof; Hartwig Schmale; Sweet taste receptor interacting protein CIB1 is a general inhibitor of InsP3-dependent Ca2+releasein vivo. Journal of Neurochemistry 2008, 106, 2249-2262, 10.1111/j.1471-4159.2008.05563.x.
- Jingsong Zhu; Stacy M. Stabler; James B. Ames; Ilia Baskakov; Mervyn J. Monteiro; Calcium binding sequences in calmyrin regulates interaction with presenilin-2. Experimental Cell Research 2004, 300, 440-454, 10.1016/j.yexcr.2004.07.020.
- Joerg Heineke; Mannix Auger-Messier; Robert N Correll; Jian Xu; Matthew J Benard; Weiping Yuan; Helmut Drexler; Leslie V Parise; Jeffery D Molkentin; CIB1 is a regulator of pathological cardiac hypertrophy. Nature Medicine 2010, 16, 872-879, 10.1038/nm.2181.
- Jr. Thomas C. Freeman; Justin L. Black; Holly G. Bray; Onur Dagliyan; Yi I. Wu; Ashutosh Tripathy; Nikolay V. Dokholyan; Tina M. Leisner; Leslie V. Parise; Identification of Novel Integrin Binding Partners for Calcium and Integrin Binding Protein 1 (CIB1): Structural and Thermodynamic Basis of CIB1 Promiscuity. Biochemistry 2013, 52, 7082-7090, 10.1021/bi400678y.
- David D. SHOCK; Ulhas P. NAIK; Julia E. BRITTAIN; Suresh K. ALAHARI; John SONDEK; Leslie V. PARISE; Calcium-dependent properties of CIB binding to the integrin αIIb cytoplasmic domain and translocation to the platelet cytoskeleton. Biochem J 1999, 342 (3), 729–735.
- Christina Spilker; Thomas Dresbach; Karl-Heinz Braunewell; Reversible Translocation and Activity-Dependent Localization of the Calcium–Myristoyl Switch Protein VILIP-1 to Different Membrane Compartments in Living Hippocampal Neurons. The Journal of Neuroscience 2002, 22, 7331-7339, 10.1523/jneurosci.22-17-07331.2002.
- Wenying Zhu; Kate E. Jarman; Noor A. Lokman; Heidi Neubauer; Lorena T. Davies; Briony L. Gliddon; Houng Taing; Paul A.B. Moretti; Martin K. Oehler; Melissa Pitman; et al.Stuart M. Pitson CIB2 Negatively Regulates Oncogenic Signaling in Ovarian Cancer via Sphingosine Kinase 1. Cancer Research 2017, 77, 4823-4834, 10.1158/0008-5472.can-17-0025.
More
