Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 8 by Conner Chen and Version 7 by Conner Chen.
Shiga toxin-producing Escherichia coli (STEC) are zoonotic Gram-negative bacteria. While raw milk cheese consumption is healthful, contamination with pathogens such as STEC can occur due to poor hygiene practices at the farm level. STEC infections cause mild to serious symptoms in humans. The raw milk cheese-making process concentrates certain milk macromolecules such as proteins and milk fat globules (MFGs), allowing the intrinsic beneficial and pathogenic microflora to continue to thrive. MFGs are surrounded by a biological membrane, the milk fat globule membrane (MFGM), which has a globally positive health effect, including inhibition of pathogen adhesion.
- STEC
- MFGM
- adhesion
- raw milk
- Milk fat globule membrane proteins
1. The Mechanism of STEC-MFG Association: What Do We Know?
1.1. General Information on Bacterial Adhesion
Bacterial adhesion is a complex process involving several factors, including: (i) surface-related properties (hydrophobicity, electrical charge, roughness, and topology); (ii) cell morphological properties (size, volume, dimension, and shape); (iii) the cell surface (chemical properties, envelope type, exposed proteins, and exopolysaccharide (EPS)); and (iv) the cell’s ability to move [1]. Adhesion is a key step for bacteria (pathogenic or not), allowing colonization and growth at a host-specific site [2]. The bacterial adhesion process consists of two phases: a non-specific phase, involving physicochemical bonds, and a specific phase involving molecular factors exposed on both host and bacterial cell surface [3]. The STEC–MFG association can be seen as a host–bacteria adhesion facilitated by the origin of the MFGM and its similarities with the membrane of intestinal cells [4][5]. In this context, both biological membranes will interact together through surface components. Various glycoconjugates are anchored on the MFGM surface and can act as ligands.
1.2. Physicochemical Interactions
Non-specific interactions have been described as the first step of adhesion, which is reversible and occurs rapidly (in the order of ~1 min) [6]. The process of initial bacterial adhesion is still not clearly understood, and physicians and microbiologists are working together to clarify the mechanisms. It is widely accepted that bacterial interaction is conducted according to the Derjaguin, Landau, Verwey, and Overbeek (DLVO) theory [7] and the extended DLVO theory [8]. The DLVO theory describes the force between charged surfaces interacting through a liquid. However, this theory may not be appropriate for modeling bacterial adhesion owing to the numerous processes involved and the influence of both biological and environmental factors (pH, ionic strength, and temperature) [9][10]. The non-specific phase of adhesion is a consequence of the balance between attractive and repulsive forces that are set up between the bacterium and the surface where it could adhere [11][12]. These forces include non-covalent interactions such as electrostatic interactions or surface charges, van der Waals forces, and Lewis acid/base interactions, as well as hydrophobic interactions [13][14]. Hydrophobic interactions and surface charges are the primary forces influencing bacterial adhesion [15].
1.2.1. Cell Surface Hydrophobicity
Bacterial cell surface hydrophobicity (CSH) is probably one of the major phenomena that governs bacterial attachment to a surface [13][14]. Hydrophobic interactions are defined as the ability of two elements of similar hydrophobicity to attract each other [16]. These forces are affected by the nature of their membrane-anchored components, including amino residues that are exposed to the extracellular environment [17]. In the context of STEC and MFGs, both are surrounded by a protein-rich membrane whose anchored surface components have polar properties (e.g., proteins and phospholipids) leading to weak hydrophobic repulsions [18][19]. Interestingly, Brisson et al. showed that the adhesion of Lactobacillus reuteri to MFGs was strain-dependent, and the more the strain was hydrophobic, the more it adhered [20].
1.2.2. Electrostatic Forces
Electrostatic forces result from the presence of a double ionic layer at the surface of a particle. The bacterial cell surface is generally negatively charged because of the carboxyl and phosphate core as well as the lipopolysaccharide (LPS) located at the surface [21]. E. coli surface charge is between −30 and −45 mV at milk pH [22][23]. While there are no published STEC-specific surface charge data generated with a modern instrument, some studies have shown that STEC isolates or reference strains are weakly negative [24][25]. Native MFGs are negatively charged due to the high phospholipid content of the outer layer of the MFGM [26]. The ζ-potential of native MFGs is close to −13 mV [27][28][29]. Furthermore, Malik et al. showed that the MFGM fraction could reach −20 mV at pH 6.5 [30]. When a negative surface meets another negative surface, repulsive forces are produced. Thus, in theory, MFGs and STEC should repel each other. However, it is important to note that the bacterial surface charge should be measured in an appropriate buffer that mimics the properties of raw milk, such as milk ultrafiltration permeate. There is a lack of recent experimental data with appropriate physicochemical conditions to assess the involvement of these forces in the association of STEC with MFGs.
1.2.3. Van der Waals Forces
Van der Waals interactions are long-range attractive forces present in both polar and non-polar molecules and come mainly from the fluctuation of the internal charge of a particle. These forces are generally attractive and result from induced dipole interactions between molecules in a colloidal particle and a substrate [31]. Attractive van der Waals forces are ubiquitous between molecules [32] and could explain part of the interaction between bacteria and the MFGM. However, van der Waals interactions in MFGM–bacteria adhesion have not been studied.
1.2.4. Lewis Acid/Base Interactions
The Lewis acid/base interaction is a polar interaction where acceptor/donor electrons enable the formation of hydrogen bonds also known as Lewis bonds [33]. This link occurs whenever ligands of strong electronegativity are associated with hydrogen. These short-range bonds are strong electrostatic interactions. Kiely and Olson showed that L. casei strains and MFGs behaved as electron donors and could mediate bonds [34][35]. However, the role of Lewis bonds in MFGM–bacteria adhesion was not fully investigated.
1.3. Specific Molecular Interactions
Bacterial molecules involved in adhesion, called adhesins, recognize specific oligosaccharide moieties or peptide residues on the surface of target cells. There are many different adhesins, including porins, complex protein structures, glycoproteins, and glycolipids. Three main types of adhesin–receptor interactions have been described: lectin–glycan; protein–protein; and hydrophobin–protein [36]. Lectins are key factors in bacterial adhesion mechanisms [37][38][39]. Lectins are adhesins that recognize glycoconjugates, the sugar epitopes generally associated with proteins or lipids. Glycoconjugates are polymeric carbohydrates composed of monosaccharides arranged in chains and preferentially present on the external leaflet either attached to lipids or proteins [40]. Commonly, the polysaccharides of glycoconjugates are referred to as the ‘glycan layer’ or ‘glycocalyx’ [41]. The glycocalyx is directly exposed to the environment, allowing interactions with other cells to facilitate cell communication, immune regulation, and adhesion [42].
A wide range of STEC isolates can be responsible for human infections, and these can be genetically different [43]. However, regardless of the strain or serogroup, STEC possess virulence factors that allow attachment to intestinal epithelial cells (IECs), and these adhesion factors are generally considered essential for infection. A large range of polysaccharides exists, but only a subset is exposed at the cell surface where they can be recognized by complementary receptors. Adhesins can be found at the distal end of bacterial pili (or fimbriae). These are bacterial extracellular appendages approximately 1 to 20 μm long and <2 to 10 nm in diameter [44]. Other adhesins are anchored directly in the biological membrane of bacteria and are referred to as afimbrial adhesins [45][46].
1.3.1. MFGM as a Decoy Receptor for STEC
Douëllou et al. showed that raw milk reduced the adhesion of two STEC strains (O157:H7 str. EDL933 and O26:H11 str. 21765) to intestinal cells in vitro and in vivo, whereas pasteurized milk did not [47]. Furthermore, Brewster and Paul showed that more than 98% of the pathogenic bacteria (including STEC) added to pasteurized or homogenized milk were recovered in the pellet after centrifugation, while less than 7% were recovered from raw milk, suggesting that processing could weaken the MFG–bacteria association [48]. Another study demonstrated that only MFGM proteins and glycoproteins inhibited E. coli adhesion in the Caco-2/HT-28 model [49]. In addition, Ross et al. suggested that the anti-infective activity of MFGM is due to the interaction of bacteria with MFGM proteins and glycoproteins rather than the interaction between MFGM and host cell receptors. In addition, modifications to MFGM surfaces such as surface roughness, zeta potential, MFG size, and phospholipid content can drastically impair the adhesive proprieties of L. fermatum [29]. The MFGM can also inhibit ETEC hemagglutination, suggesting that similar motifs are present on both membranes [50].
1.3.2. MFGM Proteins and Glycoproteins Potentially Targeted by STEC
No published studies have focused on which MFGM proteins are recognized by STEC or which adhesins are involved. However, studies have been conducted on other bacterial models (mostly beneficial). Guerin et al. used atomic force microscopy (AFM) to show that the spaCBA pili of L. rhamnosus engaged with the MFGM. Another experiment conducted by Novakovic et al. demonstrated, by blot overlay, binding of the ETEC F4ac pili to various porcine MFGM or milk proteins, including lactadherin, butyrophilin, adipophilin, acyl-CoA synthetase 3, and fatty acid-binding protein 3 [51]. An extensive literature search highlighted several MFGM proteins or glycoproteins that could interact with bacteria (Table 1). As an example, Zg16 can bind peptidoglycan [52]. Milk whey proteins such as lactoferrin, β-lactoglobulin, and α-lactalbumin can be adsorbed on the MFGM by heat treatment [53][54] and can be bound by bacteria. Glycoproteins such as mucins (MUC1 and MUC15), CD59, ECM proteins (tenascin, vitronectin), butyrophilin, prolactin-inducible protein (mPIP), CD36, and alpha1-antichymotrypsin can be bacterial lectin targets (Table 1). Among this non-exhaustive list, mucins could well be potential targets for STEC. Mucins are highly glycosylated proteins known to adhere to bacteria. Mucins constitute mucus, a secreted gel that binds the intestinal microbiota and protects the epithelium from pathogens [55][56]. Additionally, EF-Tu, a ubiquitous bacterial protein that can bind many proteins and mediate adhesion, could potentially interact with the MFGM [57].
Table 1. MFGM proteins or glycoproteins that are potentially bound by STEC.
| Bovine MFGM Components | Bacterial Components | References |
|---|---|---|
| Adipophilin * (ADPH) | F4ac (E. coli) fimbria | [51] |
| Alpha 1-antichymotrypsin (serpin) | - | [58] |
| Annexins A1, A2, A5 | LPS (lipid A), OmpB, YadC (tip adhesin of Yad fimbriae) | [59][60][61] |
| Apolipoprotein serum amyloid A protein | OmpA | [62] |
| Apolipoproteins | LPS | [63][64] |
| Butyrophilin * | F4ac (E. coli) fimbria | [4][51] |
| Calnexin | LPS, peptidoglycan | [65] |
| Cathelicidin 1 | LPS, LTA | [66] |
| CD36 * | LPS, LTA | [4][67] |
| CD5L protein | - | [68] |
| Elongation factor thermal unstable Tu (EF-Tu) |
- | [57] |
| Fatty acid-binding protein * | F4ac (E. coli) fimbria | [51] |
| Fibrinogen | Fibrinogen-binding protein (MSCRAMMs), curli | [69][70][71][72][73][74] |
| Galectin 7 | LPS | [75] |
| Gelsolin | LPS, LTA | [76] |
| Immunoglobulins | Many bacterial proteins | - |
| Integrin | Many bacterial proteins | [74][77][78][79][80][81] |
| Lactadherin * | F4ac (E. coli) fimbria | [51][82] |
| Lactoferrin | OMPs | [83] |
| Macrophage scavenger receptor | LPS, LTA | [84] |
| MUC1 *, MUC15 * | Many bacterial proteins | [85] |
| Polymeric immunoglobulin receptor (PIgR) | Ig-mediated adhesion, direction interaction via adhesin | [86][87] |
| Prolactin-inducible protein (mPIP) | - | [88][89] |
| Peptidoglycan recognition protein 1 | - | [90] |
| Protein disulfide-isomerase (PDI) | - | [91] |
| Toll-like receptor 4, 2 | Many bacterial proteins | [92][93][94][95] |
| Uromodulin | Surface layer protein A, FimH | [96][97] |
| Vimentin | Many bacterial proteins | [98] |
| Vitronectin | Many bacterial proteins | [99] |
| Zymogen granule protein 16 homolog B | LTA, peptidoglycan | [100] |
| β-lactoglobulin | Spa pili | [101][102] |
Besides proteins, a strain-specific adhesion between milk phospholipids (MPLs) and lactic acid bacteria (LAB) has been shown [106][107]. D’Incecco et al. showed that in the case of the presence of Clostridium tyrobutyricum spores in raw milk, these spores can be localized at the proximity of MFGs [108]. Like bacteria, the spore’s surface is decorated by polysaccharides and anchored extracellular appendages that mediate lectin–carbohydrate interactions [109][110]. However, the surface structure of Clostridium tyrobutyricum spores involved in the association with MFG was not identified in the study. Interestingly, further experiments conducted by D’Incecco et al. used transmission electron microscopy (TEM) to show that C. tyrobuctyricum interacted with the MFGM through an amorphous substance containing IgA [111].
Milk provides not only nutrients but also protection to newborns through immunocompetent cells, antimicrobial peptides, oligosaccharides, immunoglobulins (Igs), cytokines, growth factors, and lysosomes [112]. Bovine MFGs contain numerous immune-related proteins including proteins with bacterial binding capacities. Immune proteins are well characterized and known to recognize specific epitopes on pathogens. Immunoglobulins and immune cells in milk reflect the mother’s pathogen exposure and can provide immunity against some pathogens. Studies have shown that IgA, secreted-IgA (sIgA), and IgM are concentrated in the cream layer and can adsorb onto human [113][114] or bovine [108] MFGM surfaces. These adsorbed Igs may act as mediators of bacterial adherence to MFGs. Other studies have demonstrated the efficacy of bovine Igs against various human pathogens related to diarrhea [115][116][117]. Antibodies against pathogenic E. coli are common in samples of human milk [118][119]. Several studies have also shown that bovine colostrum contains antibodies to E. coli O157:H7 and other pathogens, regardless of whether the animals were immunized (vaccinated) or not. These antibodies can confer protection against relevant pathogens to humans [120][121][122]. Oliveira et al. showed that Igs could interact with ETEC fimbrial proteins and block adhesion to host receptors [123]. It has also been reported that K88-positive E. coli adhere to MFGs through IgA [124].
The spontaneous agglutination of MFGs in cold milk is due to the presence of immunoglobulins, called cryoglobulins [125]. Cryoglobulins are large molecules that precipitate at low temperatures (<37 °C) and disperse again on warming. Cryoglobulins are probably involved in bacterial clarification during natural creaming [126]. Immunoglobulin cell receptors are present on both the bacterial surface and MFGMs and, therefore, could act as a bridge. A generic IgG receptor is present in cold-stored MFGM preparations, but bacterial interaction has not been shown [127]. The polymeric immunoglobulin receptor (pIgR) is present on intestinal epithelial cells and facilitates the transcytosis of Igs, especially IgA, and immune complexes [87][128].
Toll-like receptors 2 and 4 (TLR2 and TLR4), which recognize foreign antigens, are present at low levels on MFGMs [103]. For example, FimH, the adhesive tip from the Type 1 fimbriae of E. coli, binds to mannose, TLR4, and CD48 [93][129]. Furthermore, TLR2 recognizes lipoteichoic acid (LTA), peptidoglycan, lipoprotein, curli, and other pathogen-associated molecular patterns (PAMPs) [94][95][130]. CD36 is a scavenger receptor that binds lipopolysaccharide (LPS) and other ligands [131]. Cathelicidins are antimicrobial peptides that can bind LPS [66]. Peptidoglycan recognition protein 1 (PRP1) is an antibacterial protein that can kill Gram-positive bacteria by binding to peptidoglycans and interfering with peptidoglycan biosynthesis [90].
2. Consequences of the STEC–MFG Association
2.1. Difficulties in Detecting STEC in Raw Milk Products
STEC detection in food matrices classically relies on four different steps: sample preparation; enrichment; detection; and confirmation by bacterial isolation. The enrichment step consists of adding an enrichment broth to the matrix to enable growth of the target bacteria. In the detection step, a genetic method is implemented to detect the presence of target bacteria by PCR screening. Finally, the confirmation step is carried out. This confirmation is based on isolation of target bacteria grown on selective media. Immunoconcentration tools using magnetic beads spiked with antibodies can also be used in this step. The ISO TS13136:2012 is the standard currently used to detect and isolate STEC belonging to O157, O26, O103, O111, and O145 serogroups and carrying eae and stx genes in food samples.
STEC detection in raw milk cheeses is particularly challenging. First, bacterial DNA is extracted from a specific volume of enrichment broth. Then, STEC target genes (eae, stx, and genes encoding one of the five somatic antigens) are detected. However, the microflora of cheese contains bacteria that carry some of the genes used to screen for STEC. For example, some non-STEC strains of E. coli (such as enteropathogenic E. coli (EPEC)) carry the eae gene or contain phages carrying the stx gene. Some other bacterial strains can also carry the stx gene, including Citrobacter freundi, Shigella spp., Acinetobacter, Aeromonas spp., Hafnia alvei, Escherichia albertii, Escherichia cloacae, and enterotoxigenic E. coli (ETEC) [132].The presence of these bacteria in raw milk cheese can lead to positive PCR results even though STEC isolates are not present in the enrichment broth (false positives). Furthermore, the performance of the methods (LOD) varies depending on the methods used and the matrices analyzed.
In general, STEC detection is more difficult in cheese than in meat. The LODs of the different detection methods are approximately 5–10 cells/g and 10–50 cells/g for bovine meat and raw milk cheese, respectively [133][134]. The presence of a richer flora and a higher amount of fat in cheese compared with meat may be an explanation. As discussed above, bacteria are preferentially found in contact with MFGs in raw milk products; however, the available detection kits perform DNA extraction on the pelleted fraction. Moreover, MFGs can interfere with DNA extraction methods by blocking spin column filters and acting as a PCR inhibitor [135][136]. Lower efficiency of bacterial DNA extraction can lead to false negative PCR results from enrichment broth samples. Several authors have described this phenomenon for various milk origins and suggest performing the extraction on both the cream and pelleted fraction. Sun et al. showed that cream harbors bacterial species that may be underestimated when skimmed milk, rather than whole milk, is used for DNA extraction [137]. Stinson et al. showed that a significant amount of human and bacterial cells remains with the cream and that bacterial DNA profiles can vary between fractions, especially for staphylococcal species [136]. The authors suggested that high-speed centrifugation may be insufficient to pellet bacterial or eukaryotic cells from milk. Furthermore, MFGs and proteins such as caseins can disrupt the interaction between immunomagnetic beads and STEC during the confirmation step. Tween 20 can be added at this stage to improve sample homogenization and block non-specific interactions [138].
Finally, STEC isolation to confirm the presence of the bacteria is also very difficult in raw milk cheese because the cheese microbiota limits STEC growth on agar plates. In addition, the different challenges encountered during cheese processing as well as the stresses suffered by STEC during the detection protocol can lead to viable but nonculturable (VBNC) isolates [139].
These studies emphasize the importance of using whole milk instead of skimmed milk for DNA extraction, but MFGs can perturb downstream applications. To improve the recovery rate of STEC in raw milk products, it seems essential to identify the nature of the STEC–MFG association in order to dissociate the two before performing the detection process. The identification of milk components involved in PCR inhibition and the improvement of DNA purification methods would allow the development of new kits to extract bacterial DNA from milk and cream. It should be noted that, despite these limitations, available DNA kits are still very effective. Quigley et al. showed that commercial kits provided very pure DNA suitable for PCR amplification from raw milk and raw milk cheese [140]. Furthermore, ISO 6887 recommends the addition of Tween 80 when enriching high-fat matrix to improve detection and bacterial isolation. However, no formal study has shown a significant effect due to the addition of Tween 80. Finally, at the enrichment temperature (37 °C or 41.5 °C), the lipids form a surface layer that may contain the desired bacterial cells and contribute to the reduction of available oxygen and thus modify the physiological state of the bacteria (Figure 2) .
Figure 2 brings together the different concepts discussed in this section.
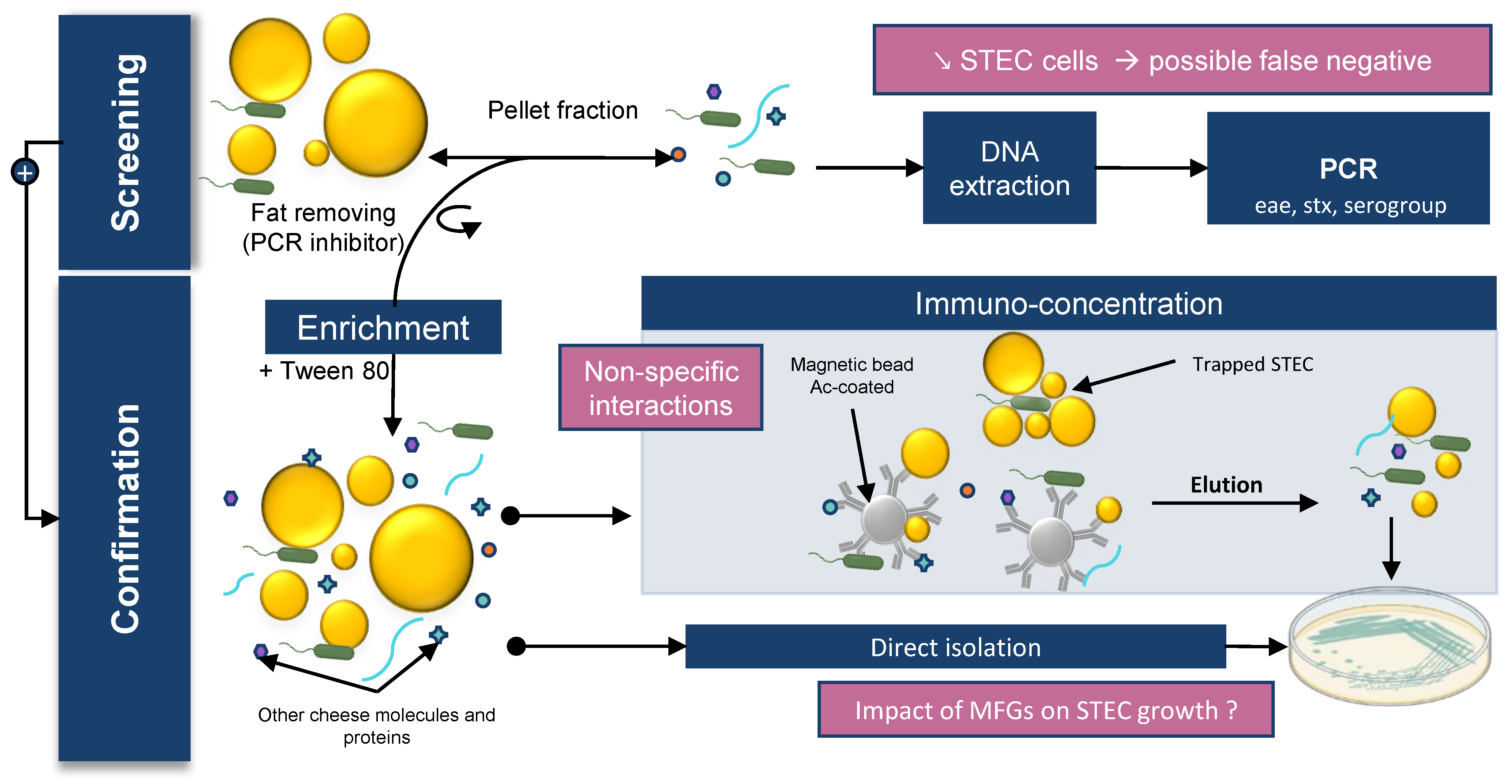
Figure 2. Impact of MFGs on STEC detection in dairy matrix. STEC detection in food matrices classically relies on 4 different steps: sample preparation; enrichment; detection; and confirmation by bacterial isolation.
2.2. Impact of Creaming on the Presence of STEC in Milk
One of the industry’s goals should be a non-invasive method to eliminate pathogenic bacteria in raw milk without affecting the nutritional qualities and raw milk microbiota of the final product. Many techniques exist, such as bactofugation and microfiltration, [141], but these techniques affect MFG structure [142] and also remove the raw milk microflora. In lab, a raw milk skimming assay was performed by electric centrifuge, and E. coli were not found in significant numbers in the cream fraction [143]. Stronger centrifugal forces are applied by the centrifuge; therefore, the STEC–MFG association is probably too weak to overcome the centrifugal forces. However, it was reported that natural creaming produces reduced-fat milk with a lower bacterial count and fewer somatic cells [144]. MFGs spontaneously rise to the surface due to the difference in density between MFGs and the aqueous phase (Stokes’ law) [145]. As previously discussed, STEC were predominantly found in the cream layer after raw milk creaming [47]. Therefore, performing natural creaming methods before cheese transformation could decrease the level of STEC in the final product. However, no study has been conducted in experimental field conditions (with low STEC contamination levels).
2.3. Anti-Adhesive Strategies
As bacterial adhesion is the first step of infection, inhibiting this step is a key strategy for infection control. Competition for the natural binding sites of pathogenic bacteria by mimetic receptors could inhibit pathogen attachment. Several natural food components could act as efficient inhibitors of pathogen adherence [2][146][147][148][149], especially milk components [4][150]. Moreover, numerous experimental studies have shown that the association of bacteria with MFGs could prevent the adhesion of several enteropathogens to enterocytes through mimetic receptors [51][82][151][152][153][154][155][156]. To avoid STEC adhesion to the epithelium of the intestinal mucosa, the STEC–MFG complex must be maintained at the site of STEC adhesion. Therefore, the expression of STEC genes involved in adhesion must be able to occur in product and during the human digestive process.
MFGM glycoconjugates are the main macromolecules involved in the anti-adhesive properties of milk against enteropathogens [4][157]. The MFGM protein fraction shows similarities to intestinal cells. Major MFGM proteins are conserved between species, although there are variations in protein expression levels and molecular functions between species and stages of lactation [158]. The MFGM was recently recognized as a high value-added ingredient, and the valorization of this by-product is constantly increasing. The MFGM, or some of its components, are added to infant milk formulas because of the MFGM’s beneficial properties [158][159][160][161][162]. More studies should be conducted to identify the MFGM surface glyco-epitopes recognized by STEC. These data could lead to pharmaceutical development of a specific drug to treat STEC infections. Currently available therapeutic solutions for STEC are controversial.
References
- Klančnik, A.; Šimunović, K.; Sterniša, M.; Ramić, D.; Smole Možina, S.; Bucar, F. Anti-Adhesion Activity of Phytochemicals to Prevent Campylobacter Jejuni Biofilm Formation on Abiotic Surfaces. Phytochem. Rev. 2020, 20, 55–84.
- Asadi, A.; Razavi, S.; Talebi, M.; Gholami, M. A Review on Anti-Adhesion Therapies of Bacterial Diseases. Infection 2019, 47, 13–23.
- Ofek, I.; Bayer, E.A.; Abraham, S.N. Bacterial Adhesion. In The Prokaryotes: Human Microbiology; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 107–123. ISBN 978-3-642-30144-5.
- Douëllou, T.; Montel, M.C.; Thevenot Sergentet, D. Invited Review: Anti-Adhesive Properties of Bovine Oligosaccharides and Bovine Milk Fat Globule Membrane-Associated Glycoconjugates against Bacterial Food Enteropathogens. J. Dairy Sci. 2017, 100, 3348–3359.
- Keenan, T.; Mather, I. Intracellular Origin of Milk Fat Globules and the Nature of the Milk Fat Globule Membrane. In Advanced Dairy Chemistry Volume 2 Lipids; Springer: Berlin/Heidelberg, Germany, 2006; pp. 137–171.
- Boks, N.P.; Kaper, H.J.; Norde, W.; Busscher, H.J.; van der Mei, H.C. Residence Time Dependent Desorption of Staphylococcus Epidermidis from Hydrophobic and Hydrophilic Substrata. Colloids Surf. B Biointerfaces 2008, 67, 276–278.
- Hermansson, M. The DLVO Theory in Microbial Adhesion. Colloids Surf. B Biointerfaces 1999, 14, 105–119.
- Bayoudh, S.; Othmane, A.; Mora, L.; Ben Ouada, H. Assessing Bacterial Adhesion Using DLVO and XDLVO Theories and the Jet Impingement Technique. Colloids Surf. B Biointerfaces 2009, 73, 1–9.
- Achinas, S.; Charalampogiannis, N.; Euverink, G.J.W. A Brief Recap of Microbial Adhesion and Biofilms. Appl. Sci. 2019, 9, 2801.
- Beloin, C.; Houry, A.; Froment, M.; Ghigo, J.-M.; Henry, N. A Short–Time Scale Colloidal System Reveals Early Bacterial Adhesion Dynamics. PLoS Biol. 2008, 6, e167.
- Katsikogianni, M.; Missirlis, Y. Concise Review of Mechanisms of Bacterial Adhesion to Biomaterials and of Techniques Used in Estimating Bacteria-Material Interactions. eCM 2004, 8, 37–57.
- Dunne, W.M. Bacterial Adhesion: Seen Any Good Biofilms Lately? Clin. Microbiol. Rev. 2002, 15, 155–166.
- van Loosdrecht, M.C.; Lyklema, J.; Norde, W.; Schraa, G.; Zehnder, A.J. The Role of Bacterial Cell Wall Hydrophobicity in Adhesion. Appl. Environ. Microbiol. 1987, 53, 1893–1897.
- Zita, A.; Hermansson, M. Determination of Bacterial Cell Surface Hydrophobicity of Single Cells in Cultures and in Wastewater in Situ. FEMS Microbiol. Lett. 1997, 152, 299–306.
- Berne, C.; Ellison, C.K.; Ducret, A.; Brun, Y.V. Bacterial Adhesion at the Single-Cell Level. Nat. Rev. Microbiol. 2018, 16, 616–627.
- Krasowska, A.; Sigler, K. How Microorganisms Use Hydrophobicity and What Does This Mean for Human Needs? Front. Cell Infect. Microbiol. 2014, 4, 112.
- Law, K.-Y. Water–Surface Interactions and Definitions for Hydrophilicity, Hydrophobicity and Superhydrophobicity. Pure Appl. Chem. 2015, 87, 759–765.
- Chevalier, F.; Sommerer, N. Analytical Methods | Mass Spectrometric Methods. In Encyclopedia of Dairy Sciences; Elsevier: Amsterdam, The Netherlands, 2011; pp. 198–205. ISBN 978-0-12-374407-4.
- Corredig, M.; Dalgleish, D.G. Effect of Heating of Cream on the Properties of Milk Fat Globule Membrane Isolates. J. Agric. Food Chem. 1998, 46, 2533–2540.
- Brisson, G.; Payken, H.F.; Sharpe, J.P.; Jiménez-Flores, R. Characterization of Lactobacillus Reuteri Interaction with Milk Fat Globule Membrane Components in Dairy Products. J. Agric. Food Chem. 2010, 58, 5612–5619.
- Hong, Y.; Brown, D.G. Electrostatic Behavior of the Charge-Regulated Bacterial Cell Surface. Langmuir 2008, 24, 5003–5009.
- Kłodzińska, E.; Szumski, M.; Dziubakiewicz, E.; Hrynkiewicz, K.; Skwarek, E.; Janusz, W.; Buszewski, B. Effect of Zeta Potential Value on Bacterial Behavior during Electrophoretic Separation. Electrophoresis 2010, 31, 1590–1596.
- Ng, W. Zeta Potential of Escherichia Coli DH5α Grown in Different Growth Media; PeerJ Inc.: Corte Madera, CA, USA, 2018.
- Lytle, D.A.; Rice, E.W.; Johnson, C.H.; Fox, K.R. Electrophoretic Mobilities of Escherichia Coli O157:H7 and Wild-Type Escherichia Coli Strains. Appl. Environ. Microbiol. 1999, 65, 3222–3225.
- Ukuku, D.O.; Fett, W.F. Relationship of Cell Surface Charge and Hydrophobicity to Strength of Attachment of Bacteria to Cantaloupe Rind†. J. Food Prot. 2002, 65, 1093–1099.
- Obeid, S.; Guyomarc’h, F.; Tanguy, G.; Leconte, N.; Rousseau, F.; Dolivet, A.; Leduc, A.; Wu, X.; Cauty, C.; Jan, G.; et al. The Adhesion of Homogenized Fat Globules to Proteins Is Increased by Milk Heat Treatment and Acidic PH: Quantitative Insights Provided by AFM Force Spectroscopy. Food Res. Int. 2019, 129, 108847.
- Michalski, M.-C.; Michel, F.; Sainmont, D.; Briard, V. Apparent ζ-Potential as a Tool to Assess Mechanical Damages to the Milk Fat Globule Membrane. Colloids Surf. B Biointerfaces 2002, 23, 23–30.
- Michalski, M.-C.; Camier, B.; Briard, V.; Leconte, N.; Gassi, J.-Y.; Goudédranche, H.; Michel, F.; Fauquant, J. The Size of Native Milk Fat Globules Affects Physico-Chemical and Functional Properties of Emmental Cheese. Le Lait 2004, 84, 343–358.
- Verma, A.; Ghosh, T.; Bhushan, B.; Packirisamy, G.; Navani, N.K.; Sarangi, P.P.; Ambatipudi, K. Characterization of Difference in Structure and Function of Fresh and Mastitic Bovine Milk Fat Globules. PLoS ONE 2019, 14, e0221830.
- Malik, P.; Danthine, S.; Paul, A.; Blecker, C. Physical-Chemical Properties of Milk Fat Globule Membrane at Different Stages of Isolation. Sci. Bull. Ser. F Biotechnol. 2015, 19, 154–159.
- Azghani, A.O.; Clark, C.A. Bacterial Infection Process: An Overview. In Regulation of the Inflammatory Response in Health and Disease; Research Signpost: Thiruvananthapuram, India, 2009; pp. 37–55. ISBN 978-81-308-0372-2.
- Kendall, K.; Roberts, A.D. Van Der Waals Forces Influencing Adhesion of Cells. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140078.
- Burgain, J.; Scher, J.; Francius, G.; Borges, F.; Corgneau, M.; Revol-Junelles, A.M.; Cailliez-Grimal, C.; Gaiani, C. Lactic Acid Bacteria in Dairy Food: Surface Characterization and Interactions with Food Matrix Components. Adv. Colloid Interface Sci. 2014, 213, 21–35.
- Kiely, L.J.; Olson, N.F. The Physicochemical Surface Characteristics of Lactobacillus Casei. Food Microbiol. 2000, 17, 277–291.
- Kiely, L.J.; Olson, N.F. Short Communication: Estimate of Non Electrostatic Interaction Free Energy Parameters for Milk Fat Globules. J. Dairy Sci. 2003, 86, 3110–3112.
- Shoaf-Sweeney, K.D.; Hutkins, R.W. Chapter 2 Adherence, Anti-Adherence, and Oligosaccharides. In Advances in Food and Nutrition Research; Elsevier: Amsterdam, The Netherlands, 2008; Volume 55, pp. 101–161. ISBN 978-0-12-374120-2.
- Cho, S.-H.; Lee, K.M.; Kim, C.-H.; Kim, S.S. Construction of a Lectin–Glycan Interaction Network from Enterohemorrhagic Escherichia Coli Strains by Multi-Omics Analysis. IJMS 2020, 21, 2681.
- Ielasi, F.S.; Alioscha-Perez, M.; Donohue, D.; Claes, S.; Sahli, H.; Schols, D.; Willaert, R.G. Lectin-Glycan Interaction Network-Based Identification of Host Receptors of Microbial Pathogenic Adhesins. mBio 2016, 7, e00584-16.
- Sharon, N.; Ofek, I. Microbial Lectins. In Comprehensive Glycoscience-From Chemistry to Systems Biology; Elsevier: Amsterdam, The Netherlands, 2007; pp. 623–659.
- Nie, S.; Cui, S.W.; Xie, M. Bioactive Polysaccharides; Academic Press: London, UK; Elsevier: San Diego, CA, USA, 2018; ISBN 978-0-12-809418-1.
- Chevalier, L.; Selim, J.; Genty, D.; Baste, J.M.; Piton, N.; Boukhalfa, I.; Hamzaoui, M.; Pareige, P.; Richard, V. Electron Microscopy Approach for the Visualization of the Epithelial and Endothelial Glycocalyx. Morphologie 2017, 101, 55–63.
- Varki, A. Biological Roles of Glycans. Glycobiology 2017, 27, 3–49.
- Monteiro, R.; Ageorges, V.; Rojas-Lopez, M.; Schmidt, H.; Weiss, A.; Bertin, Y.; Forano, E.; Jubelin, G.; Henderson, I.R.; Livrelli, V.; et al. A Secretome View of Colonisation Factors in Shiga Toxin-Encoding Escherichia Coli (STEC): From Enterohaemorrhagic E. Coli (EHEC) to Related Enteropathotypes. FEMS Microbiol. Lett. 2016, 363, fnw179.
- Dhakal, B.K.; Bower, J.M.; Mulvey, M.A.; Yang, X.H. Pili, Fimbriae☆. In Encyclopedia of Microbiology, 4th ed.; Schmidt, T.M., Ed.; Academic Press: Oxford, UK, 2019; pp. 595–613. ISBN 978-0-12-811737-8.
- McWilliams, B.D.; Torres, A.G. EHEC Adhesins. Microbiol. Spectr. 2014, 2, EHEC-0003-2013.
- Jaglic, Z.; Desvaux, M.; Weiss, A.; Nesse, L.L.; Meyer, R.L.; Demnerova, K.; Schmidt, H.; Giaouris, E.; Sipailiene, A.; Teixeira, P.; et al. Surface Adhesins and Exopolymers of Selected Foodborne Pathogens. Microbiology 2014, 160, 2561–2582.
- Douëllou, T.; Galia, W.; Kerangart, S.; Marchal, T.; Milhau, N.; Bastien, R.; Bouvier, M.; Buff, S.; Montel, M.-C.; Sergentet-Thevenot, D. Milk Fat Globules Hamper Adhesion of Enterohemorrhagic Escherichia Coli to Enterocytes: In Vitro and In Vivo Evidence. Front. Microbiol. 2018, 9, 947.
- Brewster, J.D.; Paul, M. Short Communication: Improved Method for Centrifugal Recovery of Bacteria from Raw Milk Applied to Sensitive Real-Time Quantitative PCR Detection of Salmonella spp. J. Dairy Sci. 2016, 99, 3375–3379.
- Ross, S.A.; Lane, J.A.; Kilcoyne, M.; Joshi, L.; Hickey, R.M. Defatted Bovine Milk Fat Globule Membrane Inhibits Association of Enterohaemorrhagic Escherichia Coli O157:H7 with Human HT-29 Cells. Int. Dairy J. 2016, 59, 36–43.
- Sánchez-Juanes, F.; Alonso, J.M.; Zancada, L.; Hueso, P. Glycosphingolipids from Bovine Milk and Milk Fat Globule Membranes: A Comparative Study. Adhesion to Enterotoxigenic Escherichia Coli Strains. Biol. Chem. 2009, 390, 31–40.
- Novakovic, P.; Huang, Y.Y.; Lockerbie, B.; Shahriar, F.; Kelly, J.; Gordon, J.R.; Middleton, D.M.; Loewen, M.E.; Kidney, B.A.; Simko, E. Identification of Escherichia Coli F4ac-Binding Proteins in Porcine Milk Fat Globule Membrane. Can. J. Vet. Res. 2015, 79, 120–128.
- Bergström, J.H.; Birchenough, G.M.H.; Katona, G.; Schroeder, B.O.; Schütte, A.; Ermund, A.; Johansson, M.E.V.; Hansson, G.C. Gram-Positive Bacteria Are Held at a Distance in the Colon Mucus by the Lectin-like Protein ZG16. Proc. Natl. Acad. Sci. USA 2016, 113, 13833–13838.
- Barboza, M.; Pinzon, J.; Wickramasinghe, S.; Froehlich, J.W.; Moeller, I.; Smilowitz, J.T.; Ruhaak, L.R.; Huang, J.; Lönnerdal, B.; German, J.B.; et al. Glycosylation of Human Milk Lactoferrin Exhibits Dynamic Changes During Early Lactation Enhancing Its Role in Pathogenic Bacteria-Host Interactions. Mol. Cell Proteom. 2012, 11, M111.015248.
- Ye, A.; Singh, H.; Taylor, M.; Anema, S. Interactions of Whey Proteins with Milk Fat Globule Membrane Proteins during Heat Treatment of Whole Milk. Le Lait 2004, 84, 269–283.
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin Glycan Foraging in the Human Gut Microbiome. Front. Genet. 2015, 6, 81.
- Josenhans, C.; Müthing, J.; Elling, L.; Bartfeld, S.; Schmidt, H. How Bacterial Pathogens of the Gastrointestinal Tract Use the Mucosal Glyco-Code to Harness Mucus and Microbiota: New Ways to Study an Ancient Bag of Tricks. Int. J. Med. Microbiol. 2020, 310, 151392.
- Harvey, K.L.; Jarocki, V.M.; Charles, I.G.; Djordjevic, S.P. The Diverse Functional Roles of Elongation Factor Tu (EF-Tu) in Microbial Pathogenesis. Front. Microbiol. 2019, 10, 2351.
- Bachiero, D.; Uson, S.; JimÃ, R. Lipid Binding Characterization of Lactic Acid Bacteria in Dairy Products. J. Dairy Sci. 2007, 90 (Suppl. S1), 490.
- Zhang, L.; García-Cano, I.; Jiménez-Flores, R. Characterization of Adhesion between Limosilactobacillus Reuteri and Milk Phospholipids by Density Gradient and Gene Expression. JDS Commun. 2020, 1, 29–35.
- Janganan, T.K.; Mullin, N.; Tzokov, S.B.; Stringer, S.; Fagan, R.P.; Hobbs, J.K.; Moir, A.; Bullough, P.A. Characterization of the Spore Surface and Exosporium Proteins of Clostridium Sporogenes; Implications for Clostridium Botulinum Group I Strains. Food Microbiol. 2016, 59, 205–212.
- Shuster, B.; Khemmani, M.; Nakaya, Y.; Holland, G.; Iwamoto, K.; Abe, K.; Imamura, D.; Maryn, N.; Driks, A.; Sato, T.; et al. Expansion of the Spore Surface Polysaccharide Layer in Bacillus Subtilis by Deletion of Genes Encoding Glycosyltransferases and Glucose Modification Enzymes. J. Bacteriol. 2019, 201, e00321-19.
- D’Incecco, P.; Ong, L.; Pellegrino, L.; Faoro, F.; Barbiroli, A.; Gras, S. Effect of Temperature on the Microstructure of Fat Globules and the Immunoglobulin-Mediated Interactions between Fat and Bacteria in Natural Raw Milk Creaming. J. Dairy Sci. 2018, 101, 2984–2997.
- LeBouder, E.; Rey-Nores, J.E.; Raby, A.-C.; Affolter, M.; Vidal, K.; Thornton, C.A.; Labéta, M.O. Modulation of Neonatal Microbial Recognition: TLR-Mediated Innate Immune Responses Are Specifically and Differentially Modulated by Human Milk. J. Immunol. 2006, 176, 3742–3752.
- Honkanen-Buzalski, T.; Sandholm, M. Association of Bovine Secretory Immunoglobulins with Milk Fat Globule Membranes. Comp. Immunol. Microbiol. Infect. Dis. 1981, 4, 329–342.
- Schroten, H.; Bosch, M.; Nobis-Bosch, R.; Köhler, H.; Hanisch, F.G.; Plogmann, R. Secretory Immunoglobulin A Is a Component of the Human Milk Fat Globule Membrane. Pediatr. Res. 1999, 45, 82–86.
- Korhonen, H.; Marnila, P.; Gill, H.S. Bovine Milk Antibodies for Health. Br. J. Nutr. 2000, 84, 135–146.
- Mitra, A.K.; Mahalanabis, D.; Ashraf, H.; Unicomb, L.; Eeckels, R.; Tzipori, S. Hyperimmune Cow Colostrum Reduces Diarrhoea Due to Rotavirus: A Double-Blind, Controlled Clinical Trial. Acta Paediatr. 1995, 84, 996–1001.
- Tawfeek, H.I.; Najim, N.H.; Al-Mashikhi, S. Efficacy of an Infant Formula Containing Anti-Escherichia Coli Colostral Antibodies from Hyperimmunized Cows in Preventing Diarrhea in Infants and Children: A Field Trial. Int. J. Infect. Dis. 2003, 7, 120–128.
- Adachi, E.; Tanaka, H.; Toyoda, N.; Takeda, T. Detection of bactericidal antibody in the breast milk of a mother infected with enterohemorrhagic Escherichia Coli O157:H7. Kansenshogaku Zasshi 1999, 73, 451–456.
- Noguera-Obenza, M.; Ochoa, T.J.; Gomez, H.F.; Guerrero, M.L.; Herrera-Insua, I.; Morrow, A.L.; Ruiz-Palacios, G.; Pickering, L.K.; Guzman, C.A.; Cleary, T.G. Human Milk Secretory Antibodies against Attaching and Effacing Escherichia Coli Antigens. Emerg. Infect. Dis. 2003, 9, 545–551.
- Funatogawa, K.; Ide, T.; Kirikae, F.; Saruta, K.; Nakano, M.; Kirikae, T. Use of Immunoglobulin Enriched Bovine Colostrum against Oral Challenge with Enterohaemorrhagic Escherichia Coli O157:H7 in Mice. Microbiol. Immunol. 2002, 46, 761–766.
- Funatogawa, K.; Tada, T.; Kuwahara-Arai, K.; Kirikae, T.; Takahashi, M. Enriched Bovine IgG Fraction Prevents Infections with Enterohaemorrhagic Escherichia Coli O157:H7, Salmonella Enterica Serovar Enteritidis, and Mycobacterium Avium. Food Sci. Nutr. 2019, 7, 2726–2730.
- Rabinovitz, B.C.; Gerhardt, E.; Tironi Farinati, C.; Abdala, A.; Galarza, R.; Vilte, D.A.; Ibarra, C.; Cataldi, A.; Mercado, E.C. Vaccination of Pregnant Cows with EspA, EspB, γ-Intimin, and Shiga Toxin 2 Proteins from Escherichia Coli O157:H7 Induces High Levels of Specific Colostral Antibodies That Are Transferred to Newborn Calves. J. Dairy Sci. 2012, 95, 3318–3326.
- de Oliveira, I.R.; Bessler, H.C.; Bao, S.N.; del Lima, R.; Giugliano, L.G. Inhibition of Enterotoxigenic Escherichia Coli (ETEC) Adhesion to Caco-2 Cells by Human Milk and Its Immunoglobulin and Non-Immunoglobulin Fractions. Braz. J. Microbiol. 2007, 38, 86–92.
- Atroshi, F.; Alaviuhkola, T.; Schildt, R.; Sandholm, M. Fat Globule Membrane of Sow Milk as a Target for Adhesion of K88-Positive Escherichia Coli. Comp. Immunol. Microbiol. Infect. Dis. 1983, 6, 235–245.
- Mulder, H.; Walstra, P. The Milk Fat Globule; Commonwealth Agricultural Bureaux Farnham Royal: Slough, UK, 1974; ISBN 0-85198-289-1.
- Frankel, G.; Lider, O.; Hershkoviz, R.; Mould, A.P.; Kachalsky, S.G.; Candy, D.C.A.; Cahalon, L.; Humphries, M.J.; Dougan, G. The Cell-Binding Domain of Intimin from Enteropathogenic Escherichia Coli Binds to Β1 Integrins. J. Biol. Chem. 1996, 271, 20359–20364.
- Geer, S.R.; Barbano, D.M. The Effect of Immunoglobulins and Somatic Cells on the Gravity Separation of Fat, Bacteria, and Spores in Pasteurized Whole Milk1. J. Dairy Sci. 2014, 97, 2027–2038.
- Hansen, S.F.; Larsen, L.B.; Wiking, L. Thermal Effects on IgM-Milk Fat Globule-Mediated Agglutination. J. Dairy Res. 2019, 86, 108–113.
- Johansen, F.-E.; Kaetzel, C.S. Regulation of the Polymeric Immunoglobulin Receptor and IgA Transport: New Advances in Environmental Factors That Stimulate PIgR Expression and Its Role in Mucosal Immunity. Mucosal Immunol. 2011, 4, 598–602.
- Turula, H.; Wobus, C.E. The Role of the Polymeric Immunoglobulin Receptor and Secretory Immunoglobulins during Mucosal Infection and Immunity. Viruses 2018, 10, 237.
- Mossman, K.L.; Mian, M.F.; Lauzon, N.M.; Gyles, C.L.; Lichty, B.; Mackenzie, R.; Gill, N.; Ashkar, A.A. Cutting Edge: FimH Adhesin of Type 1 Fimbriae Is a Novel TLR4 Ligand. J. Immunol. 2008, 181, 6702–6706.
- Zhang, W.; Xu, L.; Park, H.-B.; Hwang, J.; Kwak, M.; Lee, P.C.W.; Liang, G.; Zhang, X.; Xu, J.; Jin, J.-O. Escherichia Coli Adhesion Portion FimH Functions as an Adjuvant for Cancer Immunotherapy. Nat. Commun. 2020, 11, 1187.
- Oliveira-Nascimento, L.; Massari, P.; Wetzler, L.M. The Role of TLR2 in Infection and Immunity. Front. Immunol. 2012, 3, 79.
- Rapsinski, G.J.; Newman, T.N.; Oppong, G.O.; van Putten, J.P.M.; Tükel, Ç. CD14 Protein Acts as an Adaptor Molecule for the Immune Recognition of Salmonella Curli Fibers. J. Biol. Chem. 2013, 288, 14178–14188.
- Tükel, Ç.; Nishimori, J.H.; Wilson, R.P.; Winter, M.G.; Keestra, A.M.; van Putten, J.P.M.; Bäumler, A.J. Toll-like Receptors 1 and 2 Cooperatively Mediate Immune Responses to Curli, a Common Amyloid from Enterobacterial Biofilms. Cell Microbiol. 2010, 12, 1495–1505.
- Lenehan, D.; Murray, A.; Smith, S.; Uhlin, B.E.; Mitchell, J. Characterisation of E. Coli Lipopolysaccharide Adherence to Platelet Receptors. Blood 2016, 128, 4906.
- van Harten, R.M.; van Woudenbergh, E.; van Dijk, A.; Haagsman, H.P. Cathelicidins: Immunomodulatory Antimicrobials. Vaccines 2018, 6, 63.
- Lu, X.; Wang, M.; Qi, J.; Wang, H.; Li, X.; Gupta, D.; Dziarski, R. Peptidoglycan Recognition Proteins Are a New Class of Human Bactericidal Proteins. J. Biol. Chem. 2006, 281, 5895–5907.
- Bao, J.; Pan, G.; Poncz, M.; Wei, J.; Ran, M.; Zhou, Z. Serpin Functions in Host-Pathogen Interactions. PeerJ 2018, 6, e4557.
- He, X.; Zhang, W.; Chang, Q.; Su, Z.; Gong, D.; Zhou, Y.; Xiao, J.; Drelich, A.; Liu, Y.; Popov, V.; et al. A New Role for Host Annexin A2 in Establishing Bacterial Adhesion to Vascular Endothelial Cells: Lines of Evidence from Atomic Force Microscopy and an in Vivo Study. Lab. Investig. 2019, 99, 1650–1660.
- Li, X.; Pei, G.; Zhang, L.; Cao, Y.; Wang, J.; Yu, L.; Dianjun, W.; Gao, S.; Zhang, Z.-S.; Yao, Z.; et al. Compounds Targeting YadC of Uropathogenic Escherichia Coli and Its Host Receptor Annexin A2 Decrease Bacterial Colonization in Bladder. EBioMedicine 2019, 50, 23–33.
- Rand, J.H.; Wu, X.-X.; Lin, E.Y.; Griffel, A.; Gialanella, P.; McKitrick, J.C. Annexin A5 Binds to Lipopolysaccharide and Reduces Its Endotoxin Activity. mBio 2012, 3, e00292-11.
- Hari-Dass, R.; Shah, C.; Meyer, D.J.; Raynes, J.G. Serum Amyloid A Protein Binds to Outer Membrane Protein A of Gram-Negative Bacteria. J. Biol. Chem. 2005, 280, 18562–18567.
- Beck, W.H.J.; Adams, C.P.; Biglang-awa, I.M.; Patel, A.B.; Vincent, H.; Haas-Stapleton, E.J.; Weers, P.M.M. Apolipoprotein A-I Binding to Anionic Vesicles and Lipopolysaccharides: Role for Lysine Residues in Antimicrobial Properties. Biochim. Biophys. Acta 2013, 1828, 1503–1510.
- Han, Q.; Han, Y.; Wen, H.; Pang, Y.; Li, Q. Molecular Evolution of Apolipoprotein Multigene Family and the Original Functional Properties of Serum Apolipoprotein (LAL2) in Lampetra Japonica. Front. Immunol. 2020, 11, 1751.
- Huang, Y.; Hui, K.; Jin, M.; Yin, S.; Wang, W.; Ren, Q. Two Endoplasmic Reticulum Proteins (Calnexin and Calreticulin) Are Involved in Innate Immunity in Chinese Mitten Crab (Eriocheir Sinensis). Sci. Rep. 2016, 6, 27578.
- Baranova, I.N.; Kurlander, R.; Bocharov, A.V.; Vishnyakova, T.G.; Chen, Z.; Remaley, A.T.; Csako, G.; Patterson, A.P.; Eggerman, T.L. Role of Human CD36 in Bacterial Recognition, Phagocytosis and Pathogen-Induced C-Jun N-Terminal Kinase (JNK)-Mediated Signaling. J. Immunol. 2008, 181, 7147–7156.
- Martinez, V.G.; Escoda-Ferran, C.; Tadeu Simões, I.; Arai, S.; Orta Mascaró, M.; Carreras, E.; Martínez-Florensa, M.; Yelamos, J.; Miyazaki, T.; Lozano, F. The Macrophage Soluble Receptor AIM/Api6/CD5L Displays a Broad Pathogen Recognition Spectrum and Is Involved in Early Response to Microbial Aggression. Cell Mol. Immunol. 2014, 11, 343–354.
- Collins, J.; van Pijkeren, J.-P.; Svensson, L.; Claesson, M.J.; Sturme, M.; Li, Y.; Cooney, J.C.; van Sinderen, D.; Walker, A.W.; Parkhill, J.; et al. Fibrinogen-Binding and Platelet-Aggregation Activities of a Lactobacillus Salivarius Septicaemia Isolate Are Mediated by a Novel Fibrinogen-Binding Protein. Mol. Microbiol. 2012, 85, 862–877.
- Foster, T.J. The MSCRAMM Family of Cell-Wall-Anchored Surface Proteins of Gram-Positive Cocci. Trends Microbiol. 2019, 27, 927–941.
- Hymes, J.P.; Klaenhammer, T.R. Stuck in the Middle: Fibronectin-Binding Proteins in Gram-Positive Bacteria. Front. Microbiol. 2016, 7, 1504.
- Reinhardt, T.A.; Lippolis, J.D. Bovine Milk Fat Globule Membrane Proteome. J. Dairy Res. 2006, 73, 406–416.
- Ko, Y.-P.; Flick, M.J. Fibrinogen Is at the Interface of Host Defense and Pathogen Virulence in Staphylococcus Aureus Infection. Semin. Thromb Hemost 2016, 42, 408–421.
- Oh, Y.J.; Hubauer-Brenner, M.; Gruber, H.J.; Cui, Y.; Traxler, L.; Siligan, C.; Park, S.; Hinterdorfer, P. Curli Mediate Bacterial Adhesion to Fibronectin via Tensile Multiple Bonds. Sci. Rep. 2016, 6, 33909.
- Pizarro-Cerdá, J.; Cossart, P. Bacterial Adhesion and Entry into Host Cells. Cell 2006, 124, 715–727.
- Vasta, G.R. Roles of Galectins in Infection. Nat. Rev. Microbiol. 2009, 7, 424–438.
- D’Incecco, P.; Faoro, F.; Silvetti, T.; Schrader, K.; Pellegrino, L. Mechanisms of Clostridium Tyrobutyricum Removal through Natural Creaming of Milk: A Microscopy Study. J. Dairy Sci. 2015, 98, 5164–5172.
- Bucki, R.; Byfield, F.J.; Kulakowska, A.; McCormick, M.E.; Drozdowski, W.; Namiot, Z.; Hartung, T.; Janmey, P.A. Extracellular Gelsolin Binds Lipoteichoic Acid and Modulates Cellular Response to Proinflammatory Bacterial Wall Components. J. Immunol. 2008, 181, 4936–4944.
- Hoffmann, C.; Ohlsen, K.; Hauck, C.R. Integrin-Mediated Uptake of Fibronectin-Binding Bacteria. Eur. J. Cell Biol. 2011, 90, 891–896.
- Krukonis, E.S.; Isberg, R.R. Microbial Pathogens and Integrin Interactions. In Integrin-Ligand Interaction; Springer: Boston, MA, USA, 1997; pp. 175–197. ISBN 978-1-4757-4066-0.
- Scibelli, A.; Roperto, S.; Manna, L.; Pavone, L.M.; Tafuri, S.; Morte, R.D.; Staiano, N. Engagement of Integrins as a Cellular Route of Invasion by Bacterial Pathogens. Vet. J. 2007, 173, 482–491.
- Ulanova, M.; Gravelle, S.; Barnes, R. The Role of Epithelial Integrin Receptors in Recognition of Pulmonary Pathogens. J. Innate Immun. 2008, 1, 4–17.
- Shahriar, F.; Ngeleka, M.; Gordon, J.R.; Simko, E. Identification by Mass Spectroscopy of F4ac-Fimbrial-Binding Proteins in Porcine Milk and Characterization of Lactadherin as an Inhibitor of F4ac-Positive Escherichia Coli Attachment to Intestinal Villi in Vitro. Dev. Comp. Immunol. 2006, 30, 723–734.
- Vazquez-Juarez, R.C.; Romero, M.J.; Ascencio, F. Adhesive Properties of a LamB-like Outer-Membrane Protein and Its Contribution to Aeromonas Veronii Adhesion. J. Appl. Microbiol. 2004, 96, 700–708.
- Peiser, L.; Gough, P.J.; Kodama, T.; Gordon, S. Macrophage Class A Scavenger Receptor-Mediated Phagocytosis of Escherichia Coli: Role of Cell Heterogeneity, Microbial Strain, and Culture Conditions In Vitro. Infect. Immun. 2000, 68, 1953–1963.
- Naughton, J.; Duggan, G.; Bourke, B.; Clyne, M. Interaction of Microbes with Mucus and Mucins. Gut Microbes 2014, 5, 48–52.
- Iovino, F.; Engelen-Lee, J.-Y.; Brouwer, M.; van de Beek, D.; van der Ende, A.; Valls Seron, M.; Mellroth, P.; Muschiol, S.; Bergstrand, J.; Widengren, J.; et al. PIgR and PECAM-1 Bind to Pneumococcal Adhesins RrgA and PspC Mediating Bacterial Brain Invasion. J. Exp. Med. 2017, 214, 1619–1630.
- Lee, B.; Bowden, G.H.W.; Myal, Y. Identification of Mouse Submaxillary Gland Protein in Mouse Saliva and Its Binding to Mouse Oral Bacteria. Arch. Oral Biol. 2002, 47, 327–332.
- Schenkels, L.C.P.M.; Walgreen-Weterings, E.; Oomen, L.C.J.M.; Bolscher, J.G.M.; Veerman, E.C.I.; Nieuw Amerongen, A.V. In Vivo Binding of the Salivary Glycoprotein EP-GP (Identical to GCDFP-15) to Oral and Non-Oral Bacteria Detection and Identification of EP-GP Binding Species. Biol. Chem. 1997, 378, 83–88.
- Green, R.S.; Naimi, W.A.; Oliver, L.D.; O’Bier, N.; Cho, J.; Conrad, D.H.; Martin, R.K.; Marconi, R.T.; Carlyon, J.A. Binding of Host Cell Surface Protein Disulfide Isomerase by Anaplasma Phagocytophilum Asp14 Enables Pathogen Infection. mBio 2020, 11, e03141-19.
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461.
- Pak, J.; Pu, Y.; Zhang, Z.T.; Hasty, D.L.; Wu, X.R. Tamm-Horsfall Protein Binds to Type 1 Fimbriated Escherichia Coli and Prevents E. Coli from Binding to Uroplakin Ia and Ib Receptors. J. Biol. Chem. 2001, 276, 9924–9930.
- Yanagihara, S.; Kanaya, T.; Fukuda, S.; Nakato, G.; Hanazato, M.; Wu, X.-R.; Yamamoto, N.; Ohno, H. Uromodulin–SlpA Binding Dictates Lactobacillus Acidophilus Uptake by Intestinal Epithelial M Cells. Int. Immunol. 2017, 29, 357–363.
- Mak, T.N.; Brüggemann, H. Vimentin in Bacterial Infections. Cells 2016, 5, 18.
- Singh, B.; Su, Y.-C.; Riesbeck, K. Vitronectin in Bacterial Pathogenesis: A Host Protein Used in Complement Escape and Cellular Invasion. Mol. Microbiol. 2010, 78, 545–560.
- Baik, J.E.; Choe, H.-I.; Hong, S.W.; Kang, S.-S.; Ahn, K.B.; Cho, K.; Yun, C.-H.; Han, S.H. Human Salivary Proteins with Affinity to Lipoteichoic Acid of Enterococcus Faecalis. Mol. Immunol. 2016, 77, 52–59.
- Gomand, F.; Borges, F.; Guerin, J.; El-Kirat-Chatel, S.; Francius, G.; Dumas, D.; Burgain, J.; Gaiani, C. Adhesive Interactions Between Lactic Acid Bacteria and β-Lactoglobulin: Specificity and Impact on Bacterial Location in Whey Protein Isolate. Front. Microbiol. 2019, 10, 1512.
- Guerin, J.; Bacharouche, J.; Burgain, J.; Lebeer, S.; Francius, G.; Borges, F.; Scher, J.; Gaiani, C. Pili of Lactobacillus Rhamnosus GG Mediate Interaction with β-Lactoglobulin. Food Hydrocoll. 2016, 58, 35–41.
- Mather, I.H. Milk Lipids | Milk Fat Globule Membrane. In Encyclopedia of Dairy Sciences; Elsevier: Amsterdam, The Netherlands, 2011; pp. 680–690. ISBN 978-0-12-374407-4.
- Yang, M.; Cong, M.; Peng, X.; Wu, J.; Wu, R.; Liu, B.; Ye, W.; Yue, X. Quantitative Proteomic Analysis of Milk Fat Globule Membrane (MFGM) Proteins in Human and Bovine Colostrum and Mature Milk Samples through ITRAQ Labeling. Food Funct. 2016, 7, 2438–2450.
- WHO; FAO. Shiga Toxin-Producing Escherichia Coli (STEC) and Food: Attribution, Characterization, and Monitoring: Report; World Health Organization: Geneva, Switzerland, 2018.
- Li, B.; Liu, H.; Wang, W. Multiplex Real-Time PCR Assay for Detection of Escherichia Coli O157:H7 and Screening for Non-O157 Shiga Toxin-Producing E. Coli. BMC Microbiol. 2017, 17, 215.
- Miszczycha, S.D.; Ganet, S.; Duniere, L.; Rozand, C.; Loukiadis, E.; Thevenot-Sergentet, D. Novel Real-Time PCR Method to Detect Escherichia Coli O157:H7 in Raw Milk Cheese and Raw Ground Meat. J. Food Prot. 2012, 75, 1373–1381.
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of Commensal and Pathogenic Microorganisms with the Intestinal Mucosal Barrier. Nat. Rev. Microbiol. 2018, 16, 457.
- Stinson, L.F.; Ma, J.; Rea, A.; Dymock, M.; Geddes, D.T. Centrifugation Does Not Remove Bacteria from the Fat Fraction of Human Milk. Sci. Rep. 2021, 11, 572.
- Sun, L.; Dicksved, J.; Priyashantha, H.; Lundh, Å.; Johansson, M. Distribution of Bacteria between Different Milk Fractions, Investigated Using Culture-Dependent Methods and Molecular-Based and Fluorescent Microscopy Approaches. J. Appl. Microbiol. 2019, 127, 1028–1037.
- Park, J.Y.; Lim, M.-C.; Park, K.; Ok, G.; Chang, H.-J.; Lee, N.; Park, T.J.; Choi, S.-W. Detection of E. Coli O157:H7 in Food Using Automated Immunomagnetic Separation Combined with Real-Time PCR. Processes 2020, 8, 908.
- Yang, D.; Wang, Y.; Zhao, L.; Rao, L.; Liao, X. Extracellular PH Decline Introduced by High Pressure Carbon Dioxide Is a Main Factor Inducing Bacteria to Enter Viable but Non-Culturable State. Food Res. Int. 2022, 151, 110895.
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. A Comparison of Methods Used to Extract Bacterial DNA from Raw Milk and Raw Milk Cheese. J. Appl. Microbiol. 2012, 113, 96–105.
- Giffel, M.T.; Horst, H.V.D. Comparison between Bactofugation and Microfiltration Regarding Efficiency of Somatic Cell and Bacteria Removal. Bull. Int. Dairy Fed. 2004, 389, 49–53.
- Lopez, C. Focus on the Supramolecular Structure of Milk Fat in Dairy Products. Reprod. Nutr. Dev. 2005, 45, 497–511.
- Bagel, A.; Université de Lyon, Lyon, France; Douëllou, T.; Université de Lyon, Lyon, France; Sergentet, D.; Université de Lyon, Lyon, France. Distribution of Escherichia coli in bovine raw milk by creaming assay. Unpublished work. 2019.
- Anderson, J.F. The Relative Proportion Of Bacteria In Top Milk (Cream Layer) And Bottom Milk (Skim Milk), and Its Bearing On Infant Feeding. J. Infect. Dis. 1909, 6, 392–400.
- Bird, J. Cream Separation. Int. J. Dairy Technol. 1991, 44, 61–63.
- Ofek, I.; Hasty, D.L.; Sharon, N. Anti-Adhesion Therapy of Bacterial Diseases: Prospects and Problems. FEMS Immunol. Med. Microbiol. 2003, 38, 181–191.
- Kahane, I.; Ofek, I. Toward Anti-Adhesion Therapy for Microbial Diseases; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 978-1-4613-0415-9.
- Orth, K.; Krachler, A.-M. Made to Stick: Anti-Adhesion Therapy for Bacterial Infections: A Major Advantage in Targeting Adhesion Is That the Body Clears Invading Pathogens Instead of Killing Them. Microbe Mag. 2013, 8, 286–290.
- Sharon, N. Carbohydrates as Future Anti-Adhesion Drugs for Infectious Diseases. Biochim. Biophys. Acta 2006, 1760, 527–537.
- Horemans, T.; Kerstens, M.; Clais, S.; Struijs, K.; van den Abbeele, P.; Assche, T.V.; Maes, L.; Cos, P. Evaluation of the Anti-Adhesive Effect of Milk Fat Globule Membrane Glycoproteins on Helicobacter Pylori in the Human NCI-N87 Cell Line and C57BL/6 Mouse Model. Helicobacter 2012, 17, 312–318.
- Kvistgaard, A.S.; Pallesen, L.T.; Arias, C.F.; López, S.; Petersen, T.E.; Heegaard, C.W.; Rasmussen, J.T. Inhibitory Effects of Human and Bovine Milk Constituents on Rotavirus Infections. J. Dairy Sci. 2004, 87, 4088–4096.
- Martín-Sosa, S.; Martín, M.-J.; Hueso, P. The Sialylated Fraction of Milk Oligosaccharides Is Partially Responsible for Binding to Enterotoxigenic and Uropathogenic Escherichia Coli Human Strains. J. Nutr. 2002, 132, 3067–3072.
- Schroten, H.; Hanisch, F.G.; Plogmann, R.; Hacker, J.; Uhlenbruck, G.; Nobis-Bosch, R.; Wahn, V. Inhibition of Adhesion of S-Fimbriated Escherichia Coli to Buccal Epithelial Cells by Human Milk Fat Globule Membrane Components: A Novel Aspect of the Protective Function of Mucins in the Nonimmunoglobulin Fraction. Infect. Immun. 1992, 60, 2893–2899.
- Simon, P.M.; Goode, P.L.; Mobasseri, A.; Zopf, D. Inhibition of Helicobacter Pylori Binding to Gastrointestinal Epithelial Cells by Sialic Acid-Containing Oligosaccharides. Infect. Immun. 1997, 65, 750–757.
- Sprong, R.C.; Hulstein, M.F.E.; Lambers, T.T.; van der Meer, R. Sweet Buttermilk Intake Reduces Colonisation and Translocation of Listeria Monocytogenes in Rats by Inhibiting Mucosal Pathogen Adherence. Br. J. Nutr. 2012, 108, 2026–2033.
- Wang, X.; Hirmo, S.; Wadström, T.; Willén, R. Inhibition of Helicobacter Pylori Infection by Bovine Milk Glycoconjugates in a BALB/cA Mouse Model. J. Med. Microbiol. 2001, 50, 430–435.
- Jiménez-Flores, R.; Brisson, G. The Milk Fat Globule Membrane as an Ingredient: Why, How, When? Dairy Sci. Technol. 2008, 88, 5–18.
- Manoni, M.; Di Lorenzo, C.; Ottoboni, M.; Tretola, M.; Pinotti, L. Comparative Proteomics of Milk Fat Globule Membrane (MFGM) Proteome across Species and Lactation Stages and the Potentials of MFGM Fractions in Infant Formula Preparation. Foods 2020, 9, 1251.
- Hernell, O.; Timby, N.; Domellöf, M.; Lönnerdal, B. Clinical Benefits of Milk Fat Globule Membranes for Infants and Children. J. Pediatr. 2016, 173, S60–S65.
- Lee, H.; Padhi, E.; Hasegawa, Y.; Larke, J.; Parenti, M.; Wang, A.; Hernell, O.; Lönnerdal, B.; Slupsky, C. Compositional Dynamics of the Milk Fat Globule and Its Role in Infant Development. Front. Pediatr. 2018, 6, 313.
- da Silva, R.C.; Colleran, H.L.; Ibrahim, S.A. Milk Fat Globule Membrane in Infant Nutrition: A Dairy Industry Perspective. J. Dairy Res. 2021, 88, 105–116.
- Timby, N.; Domellöf, M.; Lönnerdal, B.; Hernell, O. Supplementation of Infant Formula with Bovine Milk Fat Globule Membranes12. Adv. Nutr. 2017, 8, 351–355.
More
