Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Dean Liu and Version 2 by Dean Liu.
Zebrafish (Danio rerio) is most important model organism to study different physiological and biological roles of different genes that are relevant to human. The physiological roles performed by carbonic anhydrases (CAs) and carbonic anhydrase related proteins (CARPs) is not an exception to this. The best-known function of CAs is the regulation of acid–base balance. In addition, studies performed with zebrafish, among others, have revealed important roles for these proteins in many other physiological processes, some of which had not yet been predicted in the light of previous studies and suggestions and the roles include such as pigmentation and motor coordination.
- carbonic anhydrase
- carbonic anhydrase related proteins
- acid-base balance
- ion transport
- pH regulation
- motor coordination
- zebrafish
1. Carbonic Anhydrase 2a Plays a Role in Ion Transport and Gas Exchange
Knockdown of ca2 leads to a decrease in H+ activity and increase in Na+ uptake at 96 hpf with concomitant upregulation of znhe3b and downregulation of zatp61a (H+-ATPase A-subunit expression) [25][1]. Indeed, ca2 plays a role in Na+ uptake and acid-base regulation mechanisms in zebrafish HR cells [25][1]. The ca2a enzyme was found to be present in cytoplasm and the suppression of ca2 resulted in a decrease in Na+ accumulation in H-MRCs. An in situ proximity ligation assay demonstrated a very close association of ca2a, Nhe3b, and Rhcg1 ammonia transporter in H-MRC, suggesting that the Ca2a, and Rhcg1 play a key role in Na+ uptake by forming a transport metabolon with Nhe3b [27][2]. The rates of CO2 excretion increased approximately 15-fold from 24 to 48 hpf, whereas the rates of O2 uptake increased only 6.7-fold over the same period, indicating a relative stimulation of CO2 excretion over O2 uptake. Treatment of 48 hpf larvae with the CA inhibitor acetazolamide resulted in CO2 excretion rates that were 52% of the value in control larvae, a significant difference that occurred in the absence of any effect on O2 uptake. Measurement of gas transfer rates at 48 hpf indicated that CA knockdown caused a significant inhibition of CO2 excretion compared to O2 uptake. These results suggest that between 24 hpf and 48 hpf, developing zebrafish begin to rely on ca2a to meet requirements for increased CO2 excretion [30][3].
2. Carbonic Anhydrase 2b Plays a Role in CO2 Excretion
The ca2b was first identified using the functional genomic approach from Ensembl and NCBI databases and confirmed by cloning and sequencing the cDNA [25][1]. Since CO2 production increases during early embryonic development, it was hypothesized that CA is needed for effective excretion of CO2 and the pattern of CA expression shows the progress with the increase in ca2b mRNA [30][3]. Indeed, the expression analysis using RT-qPCR showed the presence of ca2b mRNA as early as 0 hpf and the ca2b mRNA was significantly higher than the ca2a mRNA at 48 hpf [30][3]. Determination of rates of O2 consumption and CO2 excretion as a function of developmental stage showed significantly higher rates at 48 hpf, coinciding with the expression of ca2b mRNA. Contribution of CA activity to CO2 excretion was studied using acetazolamide, a CA inhibitor and the results of the study showed significantly lower CO2 excretion with no effect on O2 consumption compared to the control group larvae. In addition, knockdown of the ca which reduced the activity of ca2b compared to the control group larvae at 48 hpf also reduced the excretion of CO2 significantly compared to the control group larvae [30][3]. The result of the study suggested that ca2b is needed for the excretion of CO2 during early embryonic development.
3. Carbonic Anhydrase 6 Swim Bladder Development or Function
Carbonic anhydrase VI, a secretory form of CA was originally reported as gustin by Henkin et al. (1975) from human saliva and later in 1998 it was identified as CA VI [43,44][4][5]. Several studies in humans and in knockout mice have suggested that the CA VI may have role in taste perception and immunological function [45,46,47,48,49][6][7][8][9][10]. However, even more than after 40 years from the first report, the precise physiological role of CA VI is still not known. As a result of sequencing non-mammalian genomes, the presence of ca6 gene was reported in zebrafish. The encoded ca6 protein was found to that contain an additional pentraxin (PTX) domain at the c-terminal end [33][11]. The discovery of ca6 protein with c-terminal pentraxin domain is a unique combination among the members of CA enzyme family and as well as among the known pentraxins. The recombinant CA-PTX enzyme containing 530 amino acids showed a very high carbonate dehydratase activity. Light scattering studies showed that the ca6-PTX protein is pentameric in solution. The bioinformatic analysis suggested that the ancestral CA VI was a transmembrane protein. The exon coding for the cytoplasmic domain had been replaced by an exon coding for PTX, and later the therian lineage lost the PTX-coding, resulting in the secretory CA VI protein as we know it in mammals [33][11].
The immunochemical staining showed that the CA VI-PTX is expressed in the skin, heart, gills, and swim bladder with a very strong signal on the cell surfaces. The expression analysis of ca6 mRNA using qRT-PCR showed that the gene is prominently expressed in fins/tail, and brain and low levels of expression in the gills, kidney, teeth, skin, and spleen. The knockdown of ca6-ptx gene using morpholinos did not showed no major morphological developmental defects during the development suggesting that ca6 is not critical for the embryogenesis in zebrafish [33][11]. Interestingly, at 4-dpf the morphant embryos showed absence or deflated swim bladder [33][11]. The swim pattern analysis of 4-dpf morphant larvae showed decreased buoyancy and swam less efficiently compared to the control group larvae. Interestingly, when the gene expression was restored in the 5 dpf larvae, the swimming pattern also returned to almost normal. These results suggested that CA VI is required either for swim bladder development or swim bladder function. The presence of high levels of CA VI in fish and mammalian tissues allow the delivery of CA VI onto surface of physical barriers facing external environments (gut, skin, and gills in zebrafish; skin, saliva, milk, and respiratory tract in human/mouse). This finding is consistent with a proposed function associated with primary immune defense. The result from both of studies from humans and zebrafish suggest that both mammalian and fish CAVI enzymes are components of the innate immune system irrespective of the presence or absence of PTX domain.
4. Carbonic Anhydrase 8 Motor Coordination
Carbonic anhydrase VIII is also known as carbonic anhydrase related protein (CARP) and is one of the members of the catalytically inactive CA isoforms [13,50,51][12][13][14]. The catalytic inactivity is due to the absence of one of the three histidine residues required for the enzymatic activity. Among the catalytically inactive CA isoforms, CA VIII was the first to be reported based on its expression pattern in the mouse brain [52,53][15][16]. Subsequently, several studies were carried out mainly related to its expression in both in mice and humans [13,54,55,56][12][17][18][19]. The expression studies showed that the CARP VIII is highly expressed in the cerebellum and mainly in the cerebellar Purkinje cells also during the embryonic development, suggesting a role in cerebellar development [13,54,55,56][12][17][18][19]. The most convincing evidence for the involvement of CAARP VIII in motor coordination came from studies demonstrating the spontaneously occurring mutations in the car8/CA8 gene in waddles (wdl) mice and from the members of Iraqi and Saudi Arabian family [57,58,59][20][21][22]. The wdl mice with 19 bp deletion in car8 gene showed wobbly side-to-side ataxic movement throughout their life span [57][20]. The members of Iraqi and Saudi Arabian families with a mutation in the CA8 gene showed reduction in cerebellar volume with cerebellar ataxia and cognitive impairment, and in addition the members of Iraqi family showed quadrupedal gait [58,59][21][22].
CARP VIII is known to interact with inositol 1,4,5-trisphosphate receptor type 1 (ITPR1), an ion channel protein that regulates internal Ca2+ ion release [60][23]. The mutational analysis using yeast two-hybrid system showed that almost the entire CA domain of CARP VIII is required for ITPR1 binding and release of intracellular Ca2+ ion [51,60][14][23]. The co-evolutionary analysis of ca8 and itpr1b from zebrafish revealed that these two proteins evolved together and the ITPR1b interacts with ca8 [51,60][14][23]. The expression analysis showed that the zebrafish ca8 mRNA is strongly expressed in the central nervous system similar to humans [60][23]. Accordingly, the immunohistochemical analysis using human antibodies against the ca8 is strongly expressed in the cerebellar Purkinje cells. The expression pattern in zebrafish, both at mRNA and protein levels, suggested that the function of ca8 is conserved in zebrafish, corresponding to the function of CARP VII in mammals, and is required for both the development of cerebellum and motor coordination function [60][23]. The developmental expression analysis showed that presence of ca8 mRNA at 0 hpf, suggesting that the mRNA is of maternal origin and required for the development of zebrafish brain during early embryogenesis [34][24]. The expression of ca8 mRNA was also found to be high in the brain, heart, kidney, eye, and skin. Interestingly, the expression pattern of itpr1b mRNA was found to be similar to the expression of ca8 mRNA, confirming that the itpr1b protein interacts with ca8 and is required during early embryogenesis along with ca8 [34][24].
The knockdown of ca8 gene using gene specific translation and splice site blocking morpholinos showed abnormal changes in the head of the morphant embryos as early as 9 hpf. In addition, the ca8 morphant larvae exhibited fragile body, curved tail, small eye size, and pericardial edema. As the development progressed the defects in the morphant larvae became more prominent and showed shortened tail, curved body axis, absence of swim bladder, and otolith vesicles [34][24]. The TdT-UTP nick end labeling (TUNEL) assay showed apoptosis of the cells in the head region of ca8-morphant larvae [34][24]. The transmission electron microscopy studies showed increased neuronal cell death and the apoptotic changes included a condensed nucleus, fragmented mitochondrial profiles and debris of dead cells [34][24]. The ca8 morphants injected with as low as 6µM ca8-MOs showed ataxic movement pattern similar to humans with defective CARP VIII, suggesting the importance of ca8 gene for brain development early during the embryogenesis, and confirms its role in the motor coordination function, similar to humans [34][24].
5. Association of Carbonic Anhydrase 9 with Intracellular Acidosis
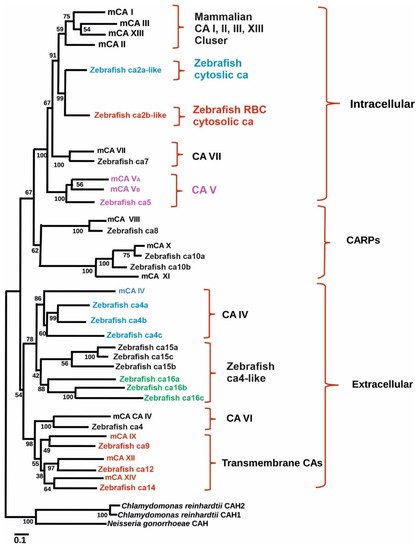
To study the role of CA IX in normal tissues in both normoxic and hypoxic environments zebrafish is an excellent model as it encounters hypoxic conditions regularly as evidenced by well-developed hypoxia responses and response systems [70,74][25][26]. Indeed, a study was conducted to examine the presence and expression of ca9 and its hypoxic response in zebrafish. Similarly, the effectiveness of hypoxia inducible CA expression in reducing intracellular acidosis was also investigated. Database search revealed the presence of hypothetical protein similar to mammalian CA IX in GenBank (XM_689890) (Figure 1). The transcript size of the putative gene contains 1560 bp with a coding region of 1155 pb. The predicted structure of the protein showed that it contains an N-terminal signal peptide (amino acids 1–22) and a single transmembrane domain (amino acids 327–349), suggesting it to be a membrane-associated CA isozyme [35][27]. Phylogenetic analyses grouped the sequence closely with mammalian CA IX isozymes (Figure 1). Interestingly, unlike NH2-terminal region of mammalian CA IX, in zebrafish there is no proteoglycan-like binding (MN) domain, suggesting that the predicted function of ca9 in hypoxia is due to the catalytic activity of the enzyme [61][28].

Expression analyses showed the presence of CA IX mRNA in the eye and gut and low levels present in the kidney, liver and brain and very low levels in muscle. There was an increase in the expression of ca9 in the brain, eye and muscle under hypoxic conditions, suggesting that ca9 shows similar hypoxic responses as previously shown in mammals. In the brain, ca9 was localized to junctions between neurons and astrocytes, and may be involved in maintaining the proper proton gradients for lactate shuttling. Genes that are transcriptionally regulated in response to hypoxia, including human CA IX, contain a gene enhancer motif termed hypoxia-responsive element (HRE), where hypoxia-inducible factor-1 (HIF-1) binds to DNA [75][29]. In zebrafish, multiple potential HREs were found within the zebrafish CA IX sequence mainly in various introns.
6. Carbonic Anhydrase 10a and 10b Is Required for Embryonic Development and Plays a Role in Motor Coordination
CARP X and CARP IX are the other two members of the catalytically inactive CA isoforms in mammals, and the catalytic inactivity is due to the absence of two and three of the possible three histidine residues, respectively, that are required for the enzymatic activity of the CAs [13][12]. The presence of CA10 and CA11 genes were first mentioned by Hewett and Tashian in 1996, based on many expressed sequence tags (ESTs) [53][16]. Later, the sequence of CA10 was discovered during the screening of cDNA library from human brain [76][30] and CA11 was identified during the construction of a physical map for the cone-rod retinal dystrophy [77][31]. The expression analysis of the CA 10 gene showed the presence of CA10 mRNA in salivary glands, kidney and brain, and the analysis in adult human and fetal tissues showed presence of significant signals in all parts of the central nervous system [76][30]. Similarly, immunohistochemical staining showed CARP X protein in many parts of the human brain [55,56][18][19]. The expression analysis of CA RP XI both at mRNA and protein levels first showed presence of CA11 mRNA only in the brain. However, a more sensitive RT-PCR method, showed positive signal in the pancreas, liver, kidney, salivary glands and spinal cord [77,78][31][32]. Immunohistochemical staining with monoclonal antibodies showed presence of CA XI protein in many parts of both adult and in fetal brain [55,78][18][32]. The expression analysis studies showed that the CA X and CA XI proteins play a pivotal role in the fetal and adult brain specimens.
Analysis of CA 10 and CA 11 sequences in multiple genomes showed that the CA11 gene emerged through a gene duplication process from CA10 after the divergence of the fish and tetrapod lineages [13][12]. As a result, the zebrafish and other ray-finned fishes have two CA10 orthologs, ca10a and ca10b [13][12]. The ca10a is highly similar to mammalian CA10 (90% identity to human CARP X at protein level), whereas ca10b is slightly more diverged with 75% identity between human CARP X and zebrafish cab. The expression analyses of ca10a and ca10b genes using RT-qPCR in zebrafish showed that these genes are strongly expressed in the nervous system and also in developing embryos. The ca10a was highly expressed in the brain, heart and eye, whereas the highest levels of ca10b were found in the ovary, brain and swim bladder, and a moderate signal was found in the testis, spleen and eye. The developmental expression showed that the ca10b mRNA was of maternal origin and with the highest presence at 0 hpf and later at 96 hpf and remained high until 168 hpf. The expression pattern of the ca10b gene suggested that this gene plays an important role in the early embryogenesis and is required throughout the developmental period in zebrafish. The presence of ca10b was also seen throughout developmental period of zebrafish embryos. The highest expression of ca10 be seen at 96 hpf and remained high till 168 hpf. The expression pattern of ca10a suggested that this gene plays an important role during the embryonic development especially after 72 hpf [36][33]. Evolutionary conservation of CA10-like genes, their ubiquitous expression pattern in different tissues, and high mRNA levels during embryonic development suggests a crucial role for CARP X-like proteins in vertebrates including zebrafish [36,51][14][33].
The morphant larvae knockdown for ca10a gene using anti-sense morpholinos showed defects in the head and had abnormal body shape and small eyes. With the progress in development, the abnormalities became more prominent showing long curved tail, and curved body, pericardial edema, and absence of swim bladder and otolith sacs. The ca10b morphant larvae injected with morpholinos developed more severe phenotypic defects compared to ca10a morphants, and the phenotypic defects appeared as early as 12 hpf. The ca10b morphant larvae had a short and abnormal shaped body already at 24 hpf, the body of the larvae was very fragile, and they showed a high mortality rate [36][33]. The ca10b morphant embryos had difficulty in hatching and showed abnormal body, smaller head and eye, mild pericardial edema and absence of otolith sacs, unutilized yolk sac and curved tail, and they could not survive beyond 3 dpf. The TUNEL assay on sections of ca10a 5 dpf morphant zebrafish larvae showed apoptotic cells especially in the head and eye regions. Similarly, large areas of apoptotic cells were observed in the head region of 5 dpf ca10b morphant larvae and a weaker signal in the tail region. The ca10a and ca10b morphant embryos showed abnormal movement pattern, suggesting the association of these genes in motor coordination function in zebrafish. The ca10a and ca10b morphant larvae could be partially rescued when ca10a and ca10b MOs were co-injected with capped human mRNAs for CA10 and CA11 genes. The partial rescue of ca10a and ca10b morphant embryos with the injection of gene-specific human mRNAs also confirmed the specificity of the ca10a and ca10b antisense morpholinos used in the study. The phenotypes of the zebrafish larvae obtained by knockdown of ca10a and ca10b using morpholinos were also confirmed by silencing these genes using CRISPR/Cas9 genome editing technology [36]. Similar to the morpholinos injected larvae, the ca10b mutated larvae showed severe phenotype with high rate of mortality at 1 dpf and larvae did not survive beyond 2 dpf. The ca10a mutated larvae showed a less severe phenotype with a lower mortality rate at 1 dpf. The expression pattern of ca10a mRNA suggests that ca10a protein plays an important role in the brain, eye and several other tissues. Accordingly, ca10b might play some roles in reproduction and brain functions.
7. Carbonic Anhydrase 14 Play an Important Role in Melanocyte Maturation
To confirm the role of ca14 on pigmentation in zebrafish, the coding region of ca14 gene was targeted using CRISPR-Cas9 system. The mutant fish showed a frameshift mutation in the third codon of ca14 gene by the deletion of two bases, and thus encoded a truncated protein. The ca14 knockout mutants showed small eye size, enlarged heart, and decreased pigmentation. The pigmentation phenotype observed in the morphant embryos was recapitulated in the genetic mutants, confirming the regulatory role of CA XIV in the maturation of melanocytes. At the adult stage, the ca14 mutation induced a visible decrease in pigmentation. The expression analysis of differentiation genes at 36 hpf, the time period when the pigment cells undergo migration and maturation showed reduced levels of tyr, tyrp1b, and dct in the mutant embryos. The down regulation of these genes confirmed that the cells are at an immature less pigmented state and that pigmentation promoting gene expression is severely reduced in the absence of ca14. Thus, the study using zebrafish as a model indicates that ca14 plays an important role in the modulation of melanocyte maturation process [37][34].
8. Carbonic Anhydrase 15a Plays a Role in Acid–Base Balance and Na+ Uptake
Knockdown of ca15a using morpholinos produced effects on the surface H+ gradient in the morphants. The ca15a morphants showed increase in the surface H+ concentration at 24 hpf, which was recovered to the normal level as observed in wild type fish at 48 hpf. Studies on the effect of ca15a knockdown on Na+ uptake function showed a significantly higher Na+ influx compared to the control and wild type embryos.
Expression analyses of relevant genes in the ca2a morphant embryos showed that there was a significant increase in the expression of ca15a, and on the other hand, the ca15a knockdown embryos showed decreased expression of zebrafish V-type proton ATPase catalytic subunit A (zatp6v1a) and increased expression of Na+/H+ exchanger 3b (zNHE3b). The findings suggested suggests that ca15a is associated with both acid–base regulation and Na+ uptake mechanism in zebrafish HR cells [25][1].
Another study was carried out to clarify the mechanism of Na+ uptake by analyzing the expression of 12 ca isoforms and the genes associated with Na+ uptake by Ito et al. [27][2]. The expression analyses showed presence of high levels ca15a mRNA in H+-ATPase/mitochondrion-rich cells (H-MRCs) in the zebrafish larvae. The expression of ca15a mRNA was dependent on salinity of the water, whereby the ca15a mRNA was upregulated at 0.03 mM NA+ water. Immunohistochemical analysis showed localization of ca15a in the apical membrane and external surface. Knockdown of ca15a using morpholinos resulted in significant reduction in Na+ accumulation in H-MRCs. Colocalization studies showed that ca15a is closely associated with Na+/H+ exchanger 3b (Nhe3b) and ammonia transporter Rhcg1. Similar to the finding of Lin et al., it showed that ca15a play a crucial role in Na+ uptake, and accordingly, ca15a probably forms a transport metabolon with Nhe3b for its physiological function [27][2].
9. Carbonic Anhydrase 15b Is Required for Migration of Primordial Germ Cells in Developing Embryos
Wang et al. carried out the expression analysis of ca15b using sequences (NM_213182) obtained from NCBI database [82][35]. The amplified cDNA sequence contained 1716 bp with a 918 bp open reading frame preceded by a 93 bp 5′-untranslated region (UTR) at N-terminal end and a 705 pb 3′ UTR. The zebrafish ca15b gene contains 9 exons and eight introns spanning 6420 bp genomic region that is located on chromosome 12. The ca15b gene encodes a 305 amino acid residue protein and contains a CA IV-XV like domain. The calculated molecular weight of the protein of the protein was 33.29 kDa, and the protein contains nine potential N-glycosylation sites and one potential O-glycosylation site. It is predicated to contain a transmembrane region in the C-terminal end of the protein (aa 280–302). The N-terminal region (aa 1–279) was predicated to be localized extracellularly and the short C-terminal tail (aa 303–305) is localized in the cytoplasm and the protein did not show any signal peptide. Phylogenetic analyses showed significant homology between ca15b is similar to the other ca15s of zebrafish and CA IV of mouse. Expression analysis using RT-PCR showed presence of ca15b mRNA in the ovary, heart, brain and muscle. In situ hybridization studies showed the presence of a mRNA signal throughout the cytoplasm of stage I to II oocytes and localized signal in the cortex of stage III oocytes and periphery of the stage IV oocytes. Western blot analysis used to study the expression of ca15b protein showed its expression specifically in the ovary, heart, brain and muscle, similar to the expression of its mRNA in different tissues. During the embryonic development, ca15b mRNA was found in every blastomere of the embryos from the one-cell stage to the blastula stages and was localized in the primordial germ cells (PGCs) from the two-cell stage to 24 hpf embryos. Based on the results of the expression analyses, it was concluded that ca15b is possibly involved in the development of PGCs and female germ cells in zebrafish.
