Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Peter Tang and Version 1 by Tasleem Arif.
Lysosomes are a critical component of the inner membrane system and are involved in various cellular biological processes, including macromolecular degradation, antigen presentation, intracellular pathogen destruction, plasma membrane repair, exosome release, cell adhesion/migration, and apoptosis. Lysosomes are a critical regulator of cellular metabolism, cancer, metastasis, and resistance to anticancer therapy. Additionally, lysosomal activities play a crucial role in acute myeloid leukemia (AML) development and progression, as well as maintaining the hematopoietic stem cells (HSCs) pool. It has been shown that AML cells undergo metabolic alterations due to chemotherapy or targeted treatment.
- AML
- lysosomes
- mitochondria
- HSCs
- NSCs
- apoptosis
1. Introduction
In 1955, Christian de Duve discovered lysosomes in hepatic tissue while investigating the cellular localization of glucose-6-phosphatase hydrolase, an enzyme thought to be connected with the mechanism of insulin action. He found that glucose-6-phosphatase hydrolase was located in “sac-like particles” that store nutrients, metabolites, and waste materials in a cell [1]. This breakthrough discovery led to the identification of several other additional extracellular and intracellular macromolecule-digesting hydrolases that were pH sensitive, such as phosphatases, nucleases, glycosidases, proteases, peptidases, sulfatases, and lipases. Collectively, these enzymes are capable of hydrolyzing almost all classes of macromolecules.
2. Biological Properties and Functions of Lysosomes
Lysosomes vary in shape, location, and function depending on the species and cell types [6][2]. Interestingly, lysosomes rapidly alter their distribution, amount, size, and activity to meet various cellular requirements. The outer lysosomal membrane is densely packed with transmembrane proteins, notably lysosome-associated membrane proteins (LAMPs), including LAMP1, LAMP2, LAMP3, LAMP4, and LAMP5. LAMP1 and LAMP2 are the most abundant lysosomal membrane proteins, contributing up to ~80% of all lysosomal membrane proteins. Other vital lysosomal membrane proteins (LMPs) include ion channels and a variety of cargo receptors, such as the Niemann–Pick C1 protein (NPC1), synaptotagmin (SYT7), chloride channel protein 7 (CLC7), and vacuolar (V)-ATPase proton (H+) pump (V-ATPase) [7][3]. At the expense of ATP hydrolysis, the V-ATPase pump transports H+ against its concentration gradient to maintain a lysosome pH between 4.5 and 5.5, which is required for the degradation of macromolecules by luminal hydrolases [8][4].
Endosomes and Golgi-derived vesicles containing lysosomal-specific hydrolytic enzymes are utilized to synthesize primary lysosomes [9][5]. The digestion of cargo is a vital function of lysosomal enzymes. The fusion of lysosomes is thought to be the primary mechanism by which internal or external cargo is degraded, and resulting metabolites are exported back to the cytoplasm for metabolic reuse or cell growth [10][6]. Lysosomes serve as metabolic signaling hubs and degradative compartments, influencing cell fate. They receive cargo from various routes, including the autophagic, endocytic, and phagocytic pathways. It is well known that lysosomes have multiple roles as degradative, clearing, and nutrition reservoirs [11][7]. Lipids, carbohydrates, proteins, nucleic acids, and damaged mitochondria are engulfed in lysosomes, where they are degraded by enzymes in the acidic lysosomal environment. The degraded products are recycled and reused in metabolic processes, stored in the lysosomal lumen for later use, or secreted by exocytosis [9,12][5][8].
The lysosomal luminal compartment acts as a hydrolytic engine, producing acidic hydrolytic enzymes such as proteases, nucleases, lipases, glycosidase phosphatases, and sulfatases. The decreased activity of these enzymes has the potential to impair cellular homeostasis. Lysosomes are vital to cellular health due to the storage of essential ions and metabolites, such as calcium, iron, and zinc, as well as hydrogen (H+), sodium, potassium (H+/Na+/K+), chloride (Cl−), and adenosine triphosphate (ATP). The amino acid (AA) levels, such as arginine and leucine, are regulated by lysosomal receptors involved in external signaling [13][9].
3. Subcellular Localization of Lysosomes
The subcellular location of lysosomes inside the cell determines their signaling characteristics. Kinesin motors of the kinesin superfamily (KIF) proteins (KIF1Bβ and KIF 2A) and a small GTP binding protein, ADP-ribosylation-factor-like GTPase 8B (ARL8B), allow lysosomes to migrate from the center of cells to the periphery and vice versa [14][10]. Environmental cues such as nutrition signaling also impact the lysosomal localization. Under nutrient-deprived conditions, lysosomes’ peripheral localization near the cell membrane suppresses mTORC1 activity and induces autophagy, whereas their perinuclear localization induces autophagy and increases interaction with mTORC1 [15][11].
Mechanistically, the FYVE-domain proteins protrudin and FYVE and coiled-coil domain autophagy adaptor 1 (FYCO1) help to guide mTOR-positive lysosomes to move towards the plasma membrane in a VPS34-dependent process [16][12]. Under serum starvation, perinuclear clustering of lysosomes inhibits the reactivation of mTORC2 [17][13]; however, under hypoxic conditions, lysosomes become dispersed, and mTORC1 activation is inhibited [18][14]. A tumor suppressor protein, folliculin (FLCN), controls the lysosome interaction with the perinuclear membrane, thus limiting lysosome location [19][15]. Importantly, lysosomal subcellular location determines the pH of the lysosomes. The acidity of the lysosomal membrane is reduced when V-ATPase activity is inhibited [20][16]. Lysosomal exocytosis, in which lysosomes fuse with the plasma membrane, requires a Ca2+ regulated process of lysosome mobility within a cell that is also engaged in various physiological activities such as plasma membrane repair and immunogenic ATP release [21][17].
4. Lysosomes as a Signaling Hub
Lysosomes, like other organelles, interact, communicate, and signal primarily at their surface to connect external signals to the cellular metabolism networks. Numerous ion channels and signaling proteins are present on the membrane of lysosomes.
- a. Growth factors and energy status
mTOR complex 1 (mTORC1), which is a vital signaling mechanism, fuels anabolic/biosynthetic pathways and inhibits catabolic processes such as autophagy. mTORC1 is regulated by a range of factors, such as energy status, growth factors, and nutrition. Tuberous sclerosis complex (TSC) is a signal complex that is composed of TSC1, TSC2, and TBC1D7 subunits. TSC negatively regulates the activity of mTORC1. TSC2 acts as a GTPase-activating protein (GAP) for Rheb and inactivates it by keeping it in a GDP-bound state. TSC2 inactivation results from either insulin or insulin-like growth factor (IGF) signaling. Activated Rheb, a protein in a GTP-bound state, activates mTORC1 [22][18]. Energy deficit cellular state induces the activation of AMP kinase at the lysosomal surface, which activates TSC2 via phosphorylation and inhibits RAPTOR activity, leading to the inhibition of mTORC1 and stimulation of catabolic pathways [22][18].
- b. Cytosolic amino acid (AA) signaling
Cytosolic AA promotes mTORC1 translocation to the lysosome surface (Figure 1), which itself is mediated by the coordinated actions of many complexes, including the Ragulator and Rag GTPases (A, B, C, and D). The Ragulator complex comprises five subunits: Lamtor1/p18, Lamtor2/p14, Lamtor3/MP1, Lamtor4/p10, and Lamtor5/HBXIP. The Lamtor/p18 subunit is anchored to the lysosomal membrane in response to AA signaling. Ragulator complex plays a role of guanine nucleotide exchange factor for Rag A/B and promotes their GTP-bound state. This translocates both the RAG GTPases and mTORC1 to the lysosomal membrane, where mTORC1 gets activated by the small GTPase Rheb [23,24][19][20].
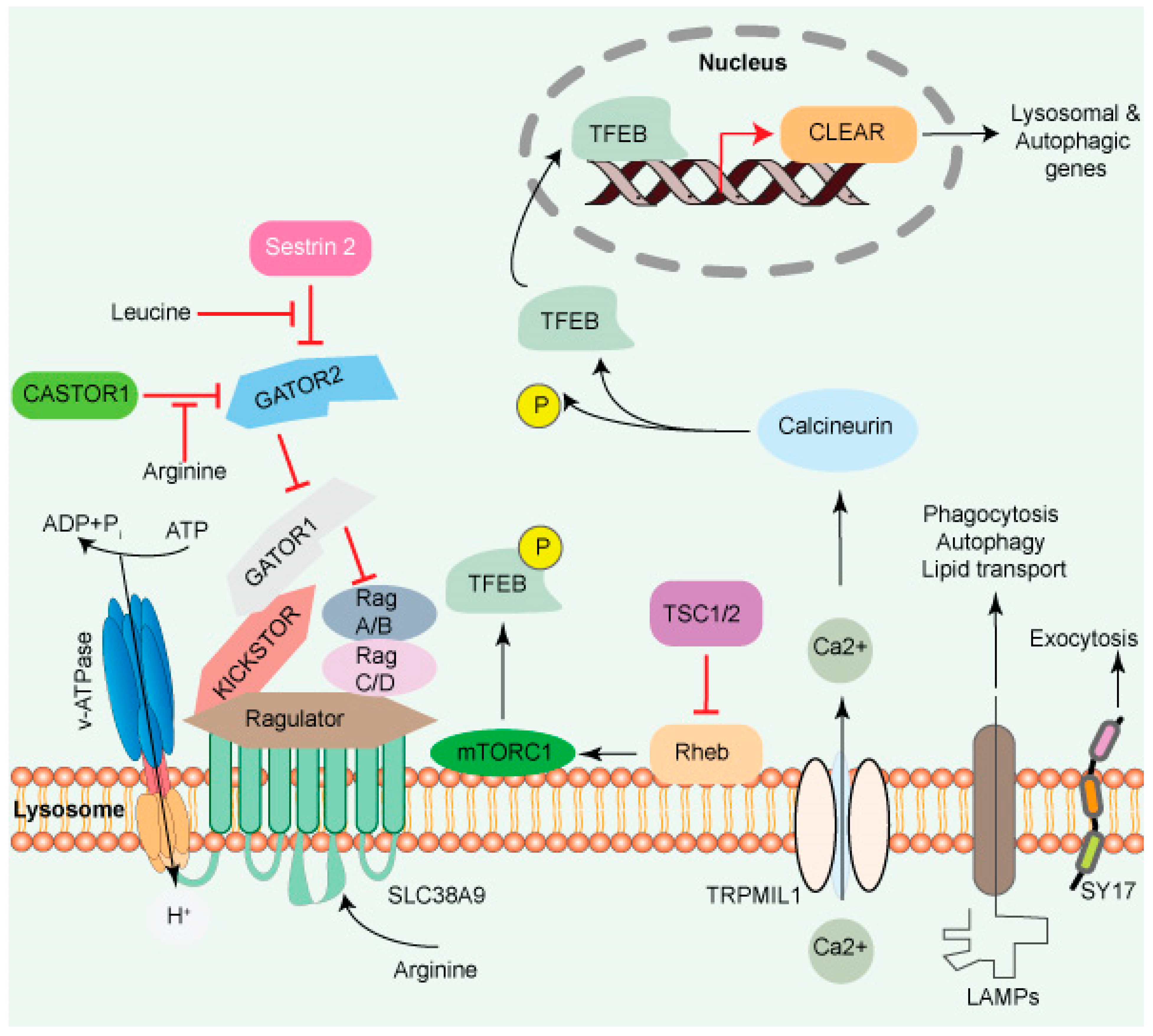

Figure 1. The lysosome surface is a hub of signaling activity. Several proteins found in lysosomal membranes are involved in various signaling cascades. The vacuolar-type H+ ATPase (V-ATPase) is a proton pump that regulates pH. mTORC1 connects metabolism and signaling. Intra- and extra-luminal amino acid signaling may affect the mTORC1 signaling pathway’s components. The amino acid transporter SLC38A9 detects arginine in the lysosomal lumen and activates mTORC1 via the Rag GTPases and Ragulator complex. A leucine sensor in the cytosol, Sestrin 2, controls mTORC1 activity with the assistance of GATOR proteins. mTORC1 also affects lysosome biogenesis adversely. Ca2+ triggers the initiation of lysosome biogenesis. Once Ca2+ is released into the cytoplasm, it dephosphorylates TFEB, allowing it to translocate to the nucleus, where it aids in the transcription of the CLEAR network genes and activates lysosomal and autophagic transcription. Transmembrane proteins such as LAMPs are involved in autophagy, lipid transport, and immunological response. SYT7 is a calcium-dependent membrane protein involved in lysosomal exocytosis.
A transporter protein, SLC38A9 facilitates the efflux of AA such as leucine from the lysosome lumen [13,28,29][9][21][22]. Increased intraluminal leucine levels also activate mTORC1 via ATP hydrolysis by the V-ATPase, which promotes the recruitment of mTORC1 by the Ragulator/Rag complex [30][23] (Figure 1). SLC38A9 in the presence of AA interacts with the Rag GTPase-Ragulator complex, which activates mTORC1 [28,29,31,32][21][22][24][25]. The tumor suppressor FLCN-FNIP complex activates RagC/D through its GAP activity. FLCN-FNIP complex, which acts as a GAP for Rag C/D, translocates to the lysosomal surface in the absence of AA [33,34][26][27]. Together, the Ragulator and FLCN/FNIP activities convert the Rag complex into its active form, which is recognized by the raptor subunit of mTORC1 [35][28], triggering its translocation to the lysosomal surfaces [36,37,38][29][30][31].
Taken together, these results suggest that lysosomes not only make a significant contribution to the cellular catabolism by providing nutrients for cell development but also serve as a platform for nutrient sensing and metabolic signal processing.
5. Regulation of Anabolic/Catabolic Pathways and mTROC1
The mTORC1 fuels anabolic/biosynthetic pathways while inhibiting catabolic processes such as autophagy (Figure 1). mTORC1 and transcription MiT/TFE transcription factor phosphorylation regulate anabolic/catabolic pathways by regulating mTORC1. The protein TFEB, which is phosphorylated on two sites in the cytoplasm, is translocated to the cytoplasm under nutrients. During stress, such as starvation, mTORC1 inhibitor induces TFEB migration to the nucleus, which initiates CLEAR network gene transcription [3][32]. Calcineurin, a Ca2+-dependent phosphatase, is another regulator of TFEB nuclear translocation. TRPML1/mucolipin-1 releases Ca2+ under starvation and triggers calcineurin, which dephosphorylates TFEB. Sixty or more genes, including microtubule-associated protein 1 light chain 3 b (Map1lc3b) and WD-repeat domain and phosphoinositide interacting 2 (Wipi2), which are involved in lysosome biogenesis and autophagy, were shown to be inhibited by a zinc finger family DNA-binding protein (ZKSCAN3). Furthermore, ZKSCAN3 and TFEB are inversely influenced by starvation [39][33]. The identification of a stress-induced lysosome-to-nucleus signaling mechanism through TFEB validates the importance of lysosomes in cellular signaling.
6. Lysosomal Intra-Luminal Compartment Signaling Events
The lysosomal luminal compartment acts as a hydrolytic engine that stores acidic hydrolytic enzymes. Any imbalance in the activity of these enzymes may have a significant effect on cellular homeostasis. Besides enzymes, lysosome also store various important ions and metabolites such as Ca2+/Fe2+/Zn2+, H+/Na+/K+, Cl−, and ATP. Additionally, lysosomal receptors control the intra-luminal levels of amino acids such as arginine and leucine and are implicated in external signaling [13][9].
- a. Lysosomes and calcium signaling
The lysosomal membrane consists of numerous ion channels that aid in establishing concentration gradients and maintaining the lysosome membrane potential [40][34]. Three distinct types of Ca2+ channels in mammalian lysosomes are reported: transient receptor potential mucolipin subfamily (TRPML)/mucolipin 1-3, two-Pore (TPC1-2), and P2X purinoceptor 4 (P2 × 4). Additionally, they respond to various cues, including cell stress, ATP depletion, phospholipids, and nutrition [41][35]. Lysosomes act as mobile intracellular Ca2+ storage, with 5000-fold higher concentrations of Ca2+ than the cytosol of the cell [42][36]. They uptake Ca2+ from the cytosol in a pH-dependent manner. Ca2+/H+ exchanger promotes lysosomal Ca2+ uptake, and recent investigations have discovered that the endoplasmic reticulum (ERs) Ca2+ levels may serve as an independent source for lysosome Ca2+ reserves [43,44,45][37][38][39].
Lysosomes contact and fuse with other organelles such as endosomes to create hybrid organelles in which the bulk of the endocytosed cargo is degraded. Various GTPases and SNARE complexes regulate them, and Ca2+ is released from the lysosome lumen [46][40]. Many research groups have reported that lysosomal processes (including mobility, trafficking, and creation of membrane contact sites) are regulated by lysosomal Ca2+ release [47,48,49,50][41][42][43][44]. The exchange of Ca2+ between the two organelles is accomplished through the ER-lysosome membrane contact sites, which are facilitated by inositol 1,4,5-trisphosphate receptors (IP3Rs), with a potential Ca2+ uptake channel/transporter in the ER/lysosome membrane [42,51][36][45]. Ca2+ concentrations in the lysosome are elevated compared to the cytoplasm, which is a primary regulator of various lysosomal functions.
These results imply that the lysosome is an essential regulatory center for several pathways involved in cell proliferation and differentiation.
- b. Essential amino acid sensing function of lysosomes
The lysosome can recognize both luminal and cytosolic amino acid (AA) levels, with the crosstalk between these pathways maintaining homeostasis or responding to nutrient-related signals. The sodium-coupled amino acid transporter SLC38A9 detects lysosomal luminal arginine. SLC38A9 undergoes a conformational change in response to arginine, which stimulates the Rag A/B (Ras-related GTP-binding) GTPase and Ragulator complex on the lysosomal surface. This activates the mechanistic target of rapamycin complex 1 (mTORC1), which is one of two protein kinase complexes incorporating the serine-threonine kinase mTOR. The mTORC1 complex consists of mTOR kinase, Raptor, GL, and DEPTOR proteins. Furthermore, mTORC1 is activated by Rheb (Ras homolog abundant in the brain) GTPase on the lysosomal membrane [52,53][46][47]. Simultaneously, SLC38A9 facilitates the efflux of other AA into the cytosol, such as leucine [13,28,29][9][21][22]. Increased intraluminal leucine levels also activate mTORC1 via ATP hydrolysis by the V-ATPase, which promotes the recruitment of mTORC1 by the Ragulator/Rag complex [30][23] (Figure 1). Taken together, these results suggest that lysosomes not only make a significant contribution to cellular catabolism, which provides nutrients for cell development, but also serve as a platform for nutrient sensing and metabolic signal processing.
- c. Lysosomal Cell Death
Lysosomal-dependent cell death (LCD) is a kind of controlled cell death that uses intra-lysosomal components such as cathepsins or iron translocation caused by lysosomal membrane permeabilization (LMP) to enhance or initiates apoptosis, autophagy, proptosis, and ferroptosis (Figure 2).
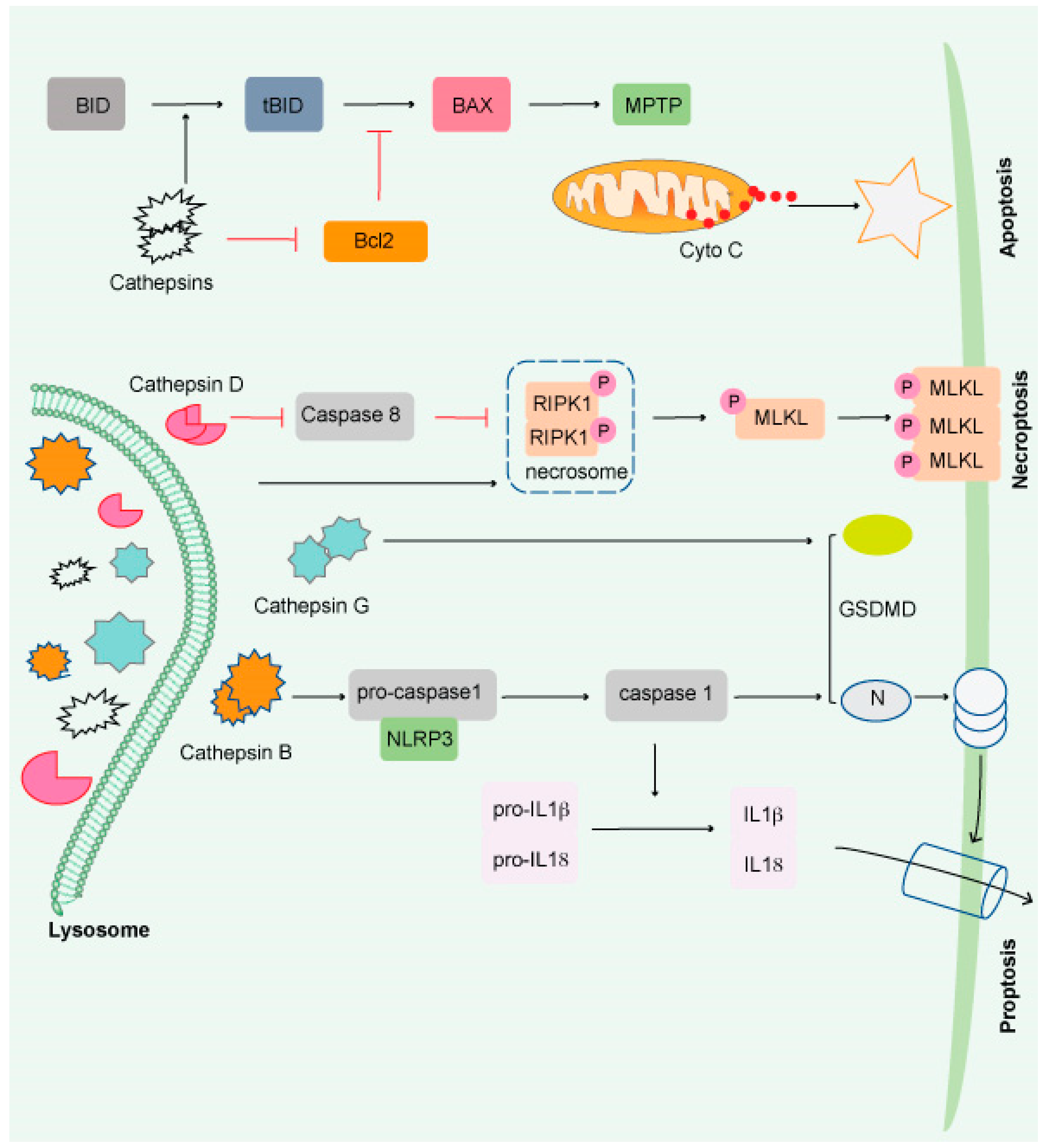

Figure 2. Impairment of the lysosome induces apoptosis. Apoptosis is triggered by lysosomal damage in a mitochondrial-dependent manner. BID is cleaved into tBid by damaged lysosomes, which increases BAX oligomorphism via inducing Bcl-2 degradation by cathepsins. As a result, BAX is translocated to the mitochondrial outer membrane (MOM), where it interacts with mitochondrial permeability y transition pore (MPTP) to release cytochrome C (Cyto C) and triggers apoptosis. Necroptosis occurs when lysosomal activity is inhibited, resulting in the accumulation of necrosome components (such as RIPK1 and RIPK3) and the release of hydrolyzed caspase 8. The necroptosis executor (MLKL) is phosphorylated and translocated to the cell membrane or organelle membrane, resulting in necrosis. Pyroptosis is induced by damaged lysosomes through cleavage of GSDMD into GSDMD-N by releasing CTSG, activation of NLRP3, and caspase-1 by the release of CTSB. Pyroptosis is induced by damaged lysosomes through the cleavage of GSDMD into GSDMD-N by the release of cathepsin G and the activation following that. Pyroptosis results in cell perforation and release of large amounts of interleukin-1 and interleukin-18.
Cathepsins (CTS), a group of lysosomal proteases that are classified as serine (CTSA and CTSG), aspartic (CTSD and E), or cysteine proteases (CTS B, C, F, H, K L, O, S, V, W, and X). Several research studies have demonstrated that most lysosomal enzymes are stable and active in a neutral and acidic pH environment and retain their degradation potential [54,55,56][48][49][50]. CTSB is the most stable protease at physiological pH and induces apoptosis [57,58,59,60][51][52][53][54]. CTSD is involved in apoptosis induced by interferon-g, Fas/CD95/APO-1, TNF-a, oxidative stress, and sphingosine [61][55]. CTSL is the least stable lysosomal protease at neutral pH and is a crucial regulator of UV-induced keratinocyte death [62,63,64][56][57][58]. The most abundant lysosomal proteases are CTSB, CTSD, and CTSL, which are found in most tissues [65][59].
Beyond protein breakdown, CTS has several other cellular functions [66][60], including cancer progression [56,67][50][61]. Extracellular CTS have been associated with matrix disintegration, cell migration, and cancer cell invasion [68][62]. Interestingly, CTS released from the lysosomal membrane induces cell death [69][63] and exhibits necrotic, apoptotic, or apoptosis-like features (Figure 2) [70][64]. Although much remains unknown, it has been shown that lysosomes play a critical role in the resistance to and initiation of cell death and the final clearance stage of cell death.
- d. Lysosomal regulation of immune responses
Lysosomes are involved in several steps of immune responses, including pathogen detection, phagocytosis, antigen processing, and inflammation. The sentinel cells, such as macrophages and dendritic cells (DCs), use toll-like receptors (TLRs) to recognize pathogen-associated molecular patterns [40][34]. Members of the TLR family, including TLR 3, TLR7, TLR 8, and TLR9, are located on endolysosomes. TLR9 senses mitochondrial DNA, which is transported to the lysosome for mitophagy, a process of removing damaged mitochondria [71,72][65][66]. When phagosomes mature, they fuse with lysosomes to form phagolysosomes, degrade foreign materials, or damaged organelles. A transcriptional factor, TFEB, increases phagocytosis in a calcium-dependent mechanism that activates immune-related genes [73][67]. Macrophages, DCs, and B cells are antigen-presenting cells (APCs) that engulf pathogens and display processed antigens on the major histocompatibility complex (MHCs) at their surfaces [74][68]. Lysosomal pH is critical for antigen processing because a highly acidic environment with low pH in the lysosomal lumen causes excessive proteolysis of the engulfed microorganism and reduces cross-presentation. On the other hand, increased lysosomal pH may affect lysosomal degradation potential and impede antigen presentation, as found in lupus disease [75][69]. In addition to their role in antigen processing, lysosomes also regulate the levels of several pro-inflammatory cytokines, such as IL-1β and IL-18, which are selectively degraded via autophagy (Figure 2) [76,77][70][71].
These results reveal that lysosomes play an essential role in modulating the intensity of the immune response and in regulating inflammation.
7. Galectins
Studies have shown that galectins mediate lysophagy, a process of removing damaged lysosomes [78][72]. Galectins play a vital role in inflammation, immunological responses, cell migration, autophagy, and signaling. Fifteen mammalian galectins (Gal-1–15) have been discovered to date [79,80][73][74]. It has been shown that apoptosis, cell cycle regulation, and nuclear splicing of pre-mRNA are all regulated by intracellular Gal-3. Many biological processes are affected by Gal-3′s interactions with various intra- and extracellular proteins under physiological and pathological conditions, such as development, immunological responses, and cancer [80][74]. Importantly, galectins have also been linked to maintaining leukemic cells in the tumor microenvironment and AML prognosis [81][75]. Extensive investigation of the lysosomal galectin may reveal their essential roles in a wide variety of biological processes, including AML, HSC biology, and other diseases.
8. Lysosomes and Autophagy
Autophagy is a process by which cells recycle their excess or damaged organelles using lysosomes, and degraded products are recycled back to the cytosol for further use. Autophagy helps cells adapt to extreme environmental conditions, such as nutritional deficiency, removing harmful/damaged organelles, destroying pathogens, and removing protein aggregates [82,83][76][77]. Autophagy is mediated by three pathways: (i) nucleotide, (ii) chaperone, and (iii) nucleotide with chaperone-dependent activities. The cargo is processed via multiple protein conjugation steps and is transported from the autophagosome to the lysosome for their degradation [84][78]. The ATG (autophagy-related) genes that are important for the autophagy process were discovered in yeast and subsequently found in other species [85][79]. The mammalian macroautophagy process begins with the formation of two major complexes: (I) the class III phosphatidylinositol 3-kinase (PI3K-III/VPS34) complex containing Beclin-1 (a mammalian homolog of yeast Atg6), hVps34, p150 (a mammalian homolog of yeast Vps15), and Atg14-like protein (Atg14L), and (II) the ULK1 kinase complex (ULK1-Atg13-FIP200-ATG101)—this activates the class III PI3K/Beclin-1 complex, enabling autophagosome nucleation formation (Figure 3). Both the above complexes are regulated by phosphorylation in the following pathways:
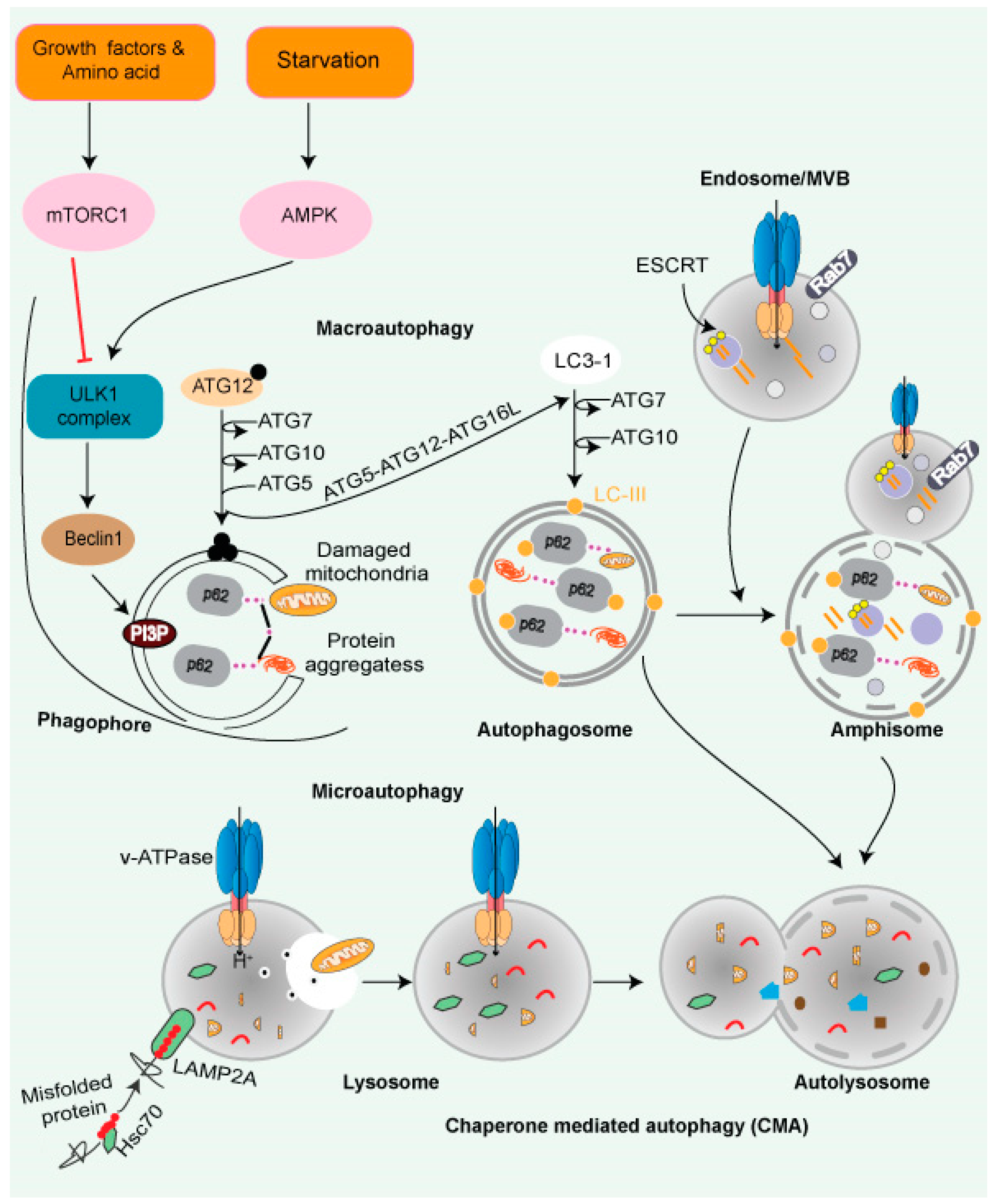

Figure 3. Autophagy and lysosomal pathway. Cytoplasmic cargo is delivered to lysosomes through autophagy. Macroautophagy includes compartmentalizing cytoplasmic payloads in LC3-coated double-membrane vesicles, known as autophagosomes. Structures known as autolysosomes are formed when autophagosomes fuse with lysosomes, and lysosomal hydrolases break down the damaged organelle. The autophagic lysosome reformation process recycles lysosomal components from autolysosomes once cargo breakdown is complete. The recognition of cytoplasmic substrates containing an accessible KFERQ-like motif by HSC70 is vital for chaperone-mediated autophagy. The HSC70-substrate complex is recognized at the lysosomal membrane by LAMP2A. LAMP2A oligomerization then forms a translocation channel, transporting the substrate into the lysosomal lumen. The LAMP2A channel disassembles into monomers after substrate delivery, allowing substrate delivery. Microautophagy entails the transport of cargo into the lysosomal lumen via interaction. Direct interaction into the endosomal lumen during endosomal microautophagy transport cargo to late endosomes, where it forms multivesicular bodies. The lysosomes then join the multivesicular bodies to degrade the cargo. Endosomal microautophagy delivers cargo via ESCRT-machinery.
- The PI3K/AKT/mTOR signaling pathway: In the abundance of nutrients and growth factors, the PI3K/AKT/mTOR signaling pathway is activated, while autophagy is suppressed and vice versa.
- AMP-activated protein kinase (AMPK): AMPK is a crucial energy sensor that monitors the ratio of ATP/AMP or ATP/ADP. Under nutrients starvation, condition-activated AMPK phosphorylates TSC2 and inactivates it, which further induces autophagy mediated by ULK-1/2 complex formation [86][80].
- Chaperone-mediated autophagy (CMA): CMA is a client-specific, selective autophagic pathway (Figure 3). CMA clients contain a signature motif containing a short stretch of 5 AA residues, namely KFERQ (lysine, phenylalanine, glutamate, arginine, glutamine) [87][81]. The HSP70 family of proteins—such as chaperone protein HSPA8, also known as HSC70 or HSP73—recognizes KFERQ [88][82]. A single molecule of LAMP-2A binds to the client protein on the lysosomal surface. Lysosomal HSP90 stabilizes multimers of LAMP-2A receptors on the lysosomal surface, facilitating the transfer of the client. Substrates are transported to the lysosome through the HSPA8-LAMP-2A interaction [89][83].
- Microautophagy is mediated by lysosome membrane dynamics, allowing cytosolic contents to be engulfed and scavenged into the lumen for degradation. Microautophagy molecular mechanisms are less understood than the other two pathways. Like CMA, the protein clients have a KFERQ-like motif and are transported to the lysosomes by HSPA8 via the endosomal sorting complex needed for the transport (ESCRT) mechanism [90][84]. The ESCRT machinery (Figure 3) helps protein migrate into multivesicular bodies (MVBs), which are a unique form of the late endosome, that can then fuse with the lysosome [91][85].
9. Role of Lysosomes in Maintaining Stem Cell Quiescence
Quiescence is required for maintaining a stem cell pool. Adult stem cells are dormant but are capable of exiting dormancy and quickly expanding and differentiating in response to stress. The quiescent state of stem cells is required for their self-renewal and is a critical factor for determining cancer stem cells’ (CSCs) susceptibility to chemotherapy and targeted treatments [92,93][86][87]. Thus, molecular underpinnings of adult stem cell quiescence are crucial for targeting quiescent CSCs in various malignancies. Recent research has increased the knowledge of the intrinsic and extrinsic regulatory mechanisms that regulate stem cell quiescence.
In the mouse brain, lysosome- and proteosome-associated gene expression was high in quiescent and active neural stem cells (NSCs), which is critical for neurogenesis [94][88].
- a. NSCs:
Quiescent NSCs (qNSCs) have a higher number of and bigger lysosomes, demonstrating compromised activity compared to active stem cells. Lysosomal gene expression is enhanced with quiescence, while their degradative capacity is reduced. These bigger lysosomes store more protein aggregates, and lysosomal activity is compromised, leading to a weak response of qNSCs to stress. On the other hand, enhancing lysosome activity reduced protein aggregates with the activation of qNSC [94][88]. Age-dependent reduction in the lysosome numbers with more protein aggregates inhibits qNSCs activation [94][88].
Interestingly, in old qNSCs, increased activity of TFEB in response to growth factors restored activation, suggesting that enhanced lysosomal function promotes activation of old NSCs. In addition, protein aggregation in lysosomes is associated with aging, which can be ameliorated by activating qNSCs [94][88]. On the contrary, a higher in vitro lysosome protease activity was reported in qNSC in vitro, with high TFEB activation and reduced NSCs growth in the adult mouse brain [95][89]. Lysosome function during quiescence and neurogenesis may differ depending on the cellular niche and NSCs’ age [96][90].
Mitf gene family members Tfe3 and Tfeb are essential for regulating lysosome biogenesis in quiescent rat embryonic fibroblasts [97][91]. The lysosome-based signaling system is a driver of mouse embryonic stem cell differentiation [98][92]. Interestingly, disrupting the distribution of lysosomal enzymes promotes the nuclear translocation of TFE3 and enhances the self-renewal of mouse embryonic stem cells (ESCs) [98][92]. MYC has an antagonistic impact on TFE3 in neoplastic cells and human iPSCs [99][93]. AMPK null embryonic stem cells suppress mTORC1 activation in the lysosome, and phosphorylation of TFE shows severe differentiation abnormalities. Due to TFEB hypophosphorylation and decreased nuclear localization, AMPK−/− ESCs retain pluripotency but fail to produce chimeric embryos. Embryoid body development requires TFEB and appropriate lysosome activity for endodermal differentiation, which it undergoes through regulation of canonical Wnt signaling [100][94]. Recent research has shown that regulating autophagy is a potential strategy for enhancing the biological characteristics of mesenchymal stem cells (MSCs) [101][95]. These results indicate that lysosome biogenesis control is essential for stem cell self-renewal or neoplastic cell self-renewal.
- b. HSCs:
Like qNSCs, quiescent hematopoietic stem cells (qHSCs) express more lysosomal genes and contain expanded lysosomes (Figure 4A) [102][96]. HSCs self-renew and remains quiescent/dormant to contribute to the expansion of the stem cells pool. When qHSCs get activated, it differentiates to produce different blood cells. qHSCs reside in a hypoxic niche of bone marrow and have modest metabolic needs, which are supplied by glycolysis [103,104][97][98]. A recent study revealed that human HSCs displayed reduced mitochondrial activity under stead-state conditions, signifying the importance of low mitochondrial activity for HSC maintenance [105][99]. HSCs actively maintain low nutrient sensitivity to maintain their quiescent non-dividing state. HSCs store and utilize nutrients to generate energy to adjust their activity, although the precise mechanism remains unclear. There has also been disagreement with the long-held belief that glycolysis is the primary energy source in quiescent HSCs (Figure 4B) [102][96]. The glycolytic pathway was more closely connected with active than quiescent HSCs under normal homeostasis, and inhibiting glycolysis in vivo enhances the potency/ quiescence of activated HSCs [102][96]. These results suggest that glycolysis is an energy source for cycling cells, which is true for embryonic stem cells and cancer cells [103][97]. Lysosomes were found to be critical in the regulation of HSC metabolism. Lysosomal degradation suppressed glycolysis and oxidative phosphorylation in the activated HSCs [102][96]. Lysosomes appear to play a critical role in maintaining the equilibrium between HSC quiescence and activation [102,106][96][100]. It has been shown that highly purified phenotypically defined HSCs have large lysosomes [102][96]. Lysosomes numbers were high in qHSCs, but few are inactivated/primed HSCs (Figure 4A,B). In addition, qHSCs had a lower lysosomal degradative potential than activated HSCs [102][96].
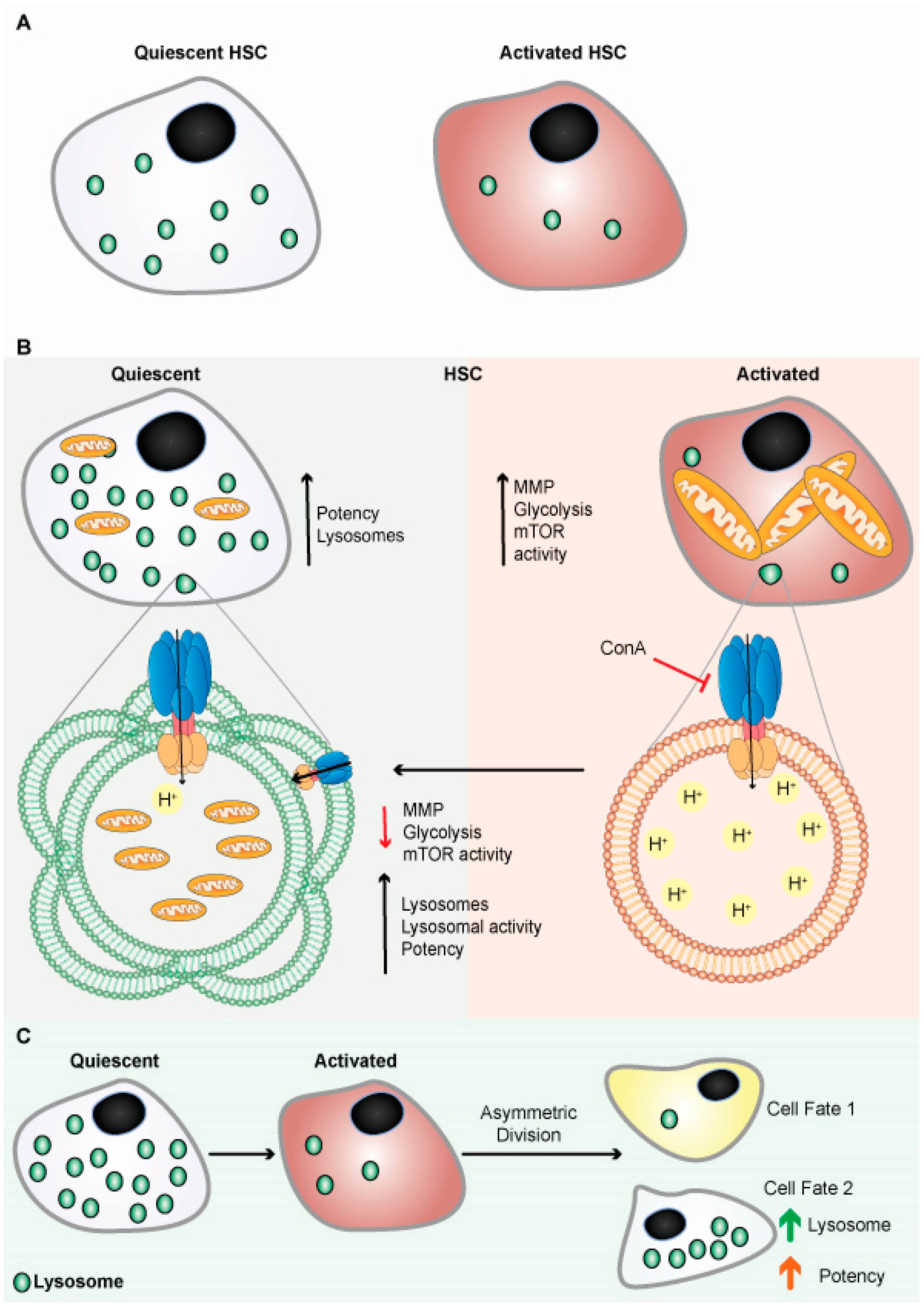

Figure 4. Lysosomes are abundant in quiescent HSCs. (A) Schematic representation shows quiescent HSCs are abundant in lysosomes. (B) Schematic representation shows quiescent HSCs are abundant in lysosomes and have poor mitochondrial lysosomal clearance. HSCs are primed by acidification and activation of lysosomes, possibly through mTORC1 activation. Lysosomes keep HSCs dormant by sequestering and storing old and defective organelles and proteins; lysosomal breakdown and release of metabolites coincide with and contribute to HSC activation and priming. (C) Lysosomes are critical for the maintenance of HSCs due to their asymmetric inheritance, which affects cell fate during cell division.
Therefore, alteration of lysosomal activity may be used to augment the potency of dormant human HSCs in transplantation settings, to combat cancer stem cells, or to improve stem cell function more broadly, including old neural stem cells, and possibly old HSCs.
