Post-translational modifications are one way that biomineral-associated cells control the function and fate of proteins. Of the ten different types of post-translational modifications, one of the most interesting and complex is glycosylation, or the covalent attachment of carbohydrates to amino acid sidechains Asn, Ser, and Thr of proteins. There are several biomineral-associated glycoproteins that have been characterized, and a subset of these have been the subject of intensive in vitro experimentation. These studies indicate that glycosylation does not alter the inherent function of the biomineralization protein; rather, it either accentuates or attenuates protein functionality. In essence, glycosylation gives the cell the “last word” as to what degree a biomineralization protein will participate in the biomineralization process.
- biomineralization
- glycosylation
- proteins
- Introduction
Over the last forty years there has been a concerted effort to understand how organisms craft biomineralized skeletal structures for survival [1][2][3][1-3]. This effort has focused along two lines: first, how do mineral crystals or amorphous minerals form under biological conditions? Recent evidence points to a mineral precursor nucleation process involving nanoparticle synthesis followed by particle assembly into larger mineral mesoscale structures [4][5][6][4-6]. Second, what agents are biosynthetically created by these same organisms to manage the mineral formation process? With regard to the latter, it has been well documented that the genomes of biomineralizing organisms code for families of proteins that are mineral-specific and unique with regard to primary sequence construction and structure [7][8][9][10][7-10]. The appearance of these proteins in the extracellular matrix during mineral formation is a clear attempt by cells to regulate the nucleation and assembly stages that lead to the final mineral product of the skeletal elements that are necessary for organism survival. Thus, to understand how biominerals form into larger, useful structures, we must understand the role or function that these proteins play in nucleation and particle assembly.
In the majority of eukaryotic organisms, the overall complexity of the biomineral proteomes is augmented by a process known as post-translational modification [11][12][13][11-13]. In essence, once a nascent protein polypeptide chain is produced on the ribosomal complexes, in some cases the cells express enzymes that perform further covalent modifications of certain amino acid sidechains on the protein, thereby altering the functionality of these sidechains. These covalent modifications occur in compartments that are separate from the cell cytoplasm [e.g., Golgi apparatus, rough endoplasmic reticulum (rER)], intracellular vesicles)[11][12][13][11-13]. A summary of common post-translational modifications (Table 1)[12] indicates that certain amino acid sidechains are targeted by cells for covalent modification. These modifications are performed by
Table 1. Common post-translational modifications of proteins.
|
Post-Translational Modification |
Type of Modification |
Site(s) of Modification |
|
Disulfide bond formation |
Thiol oxidation to form -S-S- bond |
Cys |
|
Hydroxylation |
Addition of -OH group |
Pro, Glu |
|
Ubiquitination |
Addition of ubiquitin protein(s) |
Lysine |
|
Lipidation |
Esterification to lipid group |
Cys, Lys, N-terminal Gly |
|
SUMOylation |
Small Ubiquitin-like Modifier protein |
Lys |
|
Acetylation |
Addition of acetyl group |
N-terminus, Lys, Ser |
|
Methylation |
Addition of methyl group |
Lys, Arg, termini |
|
Phosphorylation |
Addition of phosphate group |
Ser, Thr, Tyr |
|
Glycosylation |
Addition of carbohydrate group(s) |
Asn, Thr, Ser |
|
Nitration |
Addition of nitrogen |
Tyr |
|
Acylation |
Addition of acyl chain |
Cys, Gly, Ser, Thr, Lys |
intracellular enzymes and in the majority of cases the finalized protein product is transported to its intended destination using vesicles [11][12][13][11-13]. In essence, genomic information is translated into proteomic information, and in some instances, this proteomic information can further be modified and thus diversified in terms of structure and ultimately function [11][12][13][11-13].
Perhaps the most complex post-translational modification process is glycosylation, or the addition of one or more carbohydrate monomers (known as monosaccharides) to specific amino acid sidechains on a protein, thus converting the polypeptide into a glycoprotein [11][12][13][14][15][16][11-16]. There are three classifications of glycoproteins, depending on which amino acids serve as attachment points for carbohydrates [11][12][13][14][15][16][11-16]: 1) O-linked, where the oligosaccharide attachment occurs on Ser and/or Thr residues and is performed in the Golgi apparatus; 2) N-linked, where the oligosaccharide attachment occurs on Asn and is performed within the endoplasmic reticulum (ER); and 3) hybrid, in which a glycoprotein has O-linked (Ser, Thr) glycans and N-linked (Asn) glycans.
Several features contribute to the overall complexity of glycosylation [11][12][13][14][15][16][11-16]: a) The number of carbohydrate groups added to a single amino acid sidechain site can vary; b) the number and type of amino acid sites for attachment on a given protein can vary; c) at a given attachment point on a protein, the carbohydrate groups can be constructed as linear or branching chains; d) the hydroxyl-rich carbohydrate groups themselves can be modified by the addition of chemical groups, such as carboxylate, sulfate, N-acetyl amino, and hydroxyl. Thus, unlike other post-translational modifications, glycosylation represents a unique opportunity for the cell to combine two very different macromolecular building blocks (amino acids, carbohydrates) into one macromolecule, which in turn may have a significant impact on the function and distribution of this protein class within a biomineralizing system.
2. Glycoproteins in biomineralization: An overview
Table 2 provides a summary of specific mineral matrix proteins that have been identified as glycoproteins and report the complete amino acid sequence [17][18][19][20][20][21][22][23][24][25][26][27][28][29][30][17-30]. Admittedly, this table is sparse, and at the time of this writing very few biomineral-associated glycoproteins have complete protein sequence data or oligosaccharide composition/sequence data available. Note that some studies have identified glycoproteins in the extracellular matrices of different organisms [31][32][33][34][35][31-35], but to date these proteins have not been sequenced nor rigorously characterized. The majority of the identified biomineralization glycoproteins are found in association with calcium-based biominerals [17][18][19][20][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][17-35]; however, it should be acknowledged that glycoproteins may eventually be identified in other non-calcium based biominerals, such as magnetite (Fe3O4)[36] or silicates (SiO4)[37].
Table 2. Biomineral-associated glycoproteins.
|
Protein |
Organism |
Tissue |
Associated Mineral Phase |
|
SpSM30 A-F |
S. purpuratus (sea urchin) |
Embryonic spicule |
Magnesium Calcite (CaMgCO3) |
|
AP24 |
H. rufescens (abalone) |
Shell nacre |
Aragonite (CaCO3) |
|
Enamelin |
Vertebrates |
Tooth enamel |
Hydroxyapatite (CaPO4) |
|
SIBLING Family |
Vertebrates |
Bone, tooth dentine |
Hydroxyapatite (CaPO4) |
|
EDIL3 |
Avian |
Eggshell |
Calcite (CaCO3) |
|
MFGE8 |
Avian |
Eggshell |
Calcite (CaCO3) |
|
Proteoglycans |
Vertebrates |
Bone, tooth dentine |
Hydroxyapatite (CaPO4) |
3. The impact of glycosylation on protein function
Does the attachment of oligosaccharides affect the molecular behavior of a polypeptide chain? To answer this question, one could envision a comparative study wherein the function of an unglycosylated variant of a given protein is contrasted against that of a glycosylated variant, with each possessing the identical primary sequence. Here, the only variable would be the presence (or absence) of oligosaccharide chains.
Recently, this type of study was executed on two proteins, AP24 (aragonite nacre layer, Pacific red abalone H. rufescens [25] [25] and SpSM30B/C (calcitic spicule matrix, S. purpuratus, purple sea urchin)[24][28][29][30][24]. Both proteins have been the subject of in vitro glycosylation studies in insect cells, where it was discovered that AP24 and SpSM30B/C belong to the hybrid classification, i.e., they consist of N- and O-linked linear and branching oligosaccharide chains [26][27][26,27]. Interestingly, the glycosylated variants of AP24 and SpSM30B/C both contain anionic monosialylated, bisialylated and monosulfated, bisulfated monosaccharides [26][27][26,27]. Given that both proteins inhabit a Ca(II)-rich environment in vivo, the anionic monosaccharides could serve as putative sites for Ca(II)-protein or mineral-protein interactions. To a certain extent, both proteins are similar in function: they are involved in the formation of the organic matrix, forming hydrogel particles that assemble mineral nanoparticles [26][27][26,27]. In addition, both protein hydrogels become occluded within calcium carbonates and modify the material and surface properties of the minerals they inhabit [26][27][26,27]. From these two studies we note a trend where glycosylation does not change the intrinsic function of the polypeptide chain; rather, the attachment of anionic oligosaccharide moieties either 1) attenuates specific functions or has no effect (AP24); or, 2) accentuates protein functionality [SpSM30B/C]. Other studies with multiple glycoproteins will hopefully confirm this trend or provide evidence of other effects that oligosaccharides impose upon polypeptides.
In addition to modulating the mineral formation process, a key role of biomineralization proteins is the assembly and organization of multiple proteins to form an organic matrix within which the nucleation and crystal growth processes take place [1,2,7]. Studies were conducted on molluscan (AP7, AP24, H. rufescens)[28] [6,25,28] and sea urchin (SpSM50, SpSM30B/C, S. purpuratus)[29][30][6,24,29,30] recombinant two-protein systems. Here, each pair of proteins is known to co-exist in vivo within the extracellular matrix [24][6,24]. Interesting contrasts were noted: a) AP7 and AP24 protein complexes form as a direct result of polypeptide – polypeptide chain recognition and not polypeptide – oligosaccharide recognition. However, the presence of anionic oligosaccharides on AP24 appears to modulate the intensity of AP7 – AP24 protein - protein interactions and potentially stabilizes the AP24 conformation upon binding to AP7. (b) The formation of a SpSM50 – SpSM30B/C complex requires glycosylation and, in contrast to the AP7 - AP24 study described above, these interactions were found to be Ca(II) – independent for both variants [29][30][29,30]. The glycosylation requirement clearly indicates that SpSM50 polypeptide sequence recognizes and binds to the glycan moieties on the surface of SpSM30B/C.Notably, the SpSM50 protein possesses a glycan-binding motif known as the C-type lectin-like domain, and it is believed that this region interacts with the glycan groups of SpSM30 (Figure 1)[29][30][6,24,29,30].
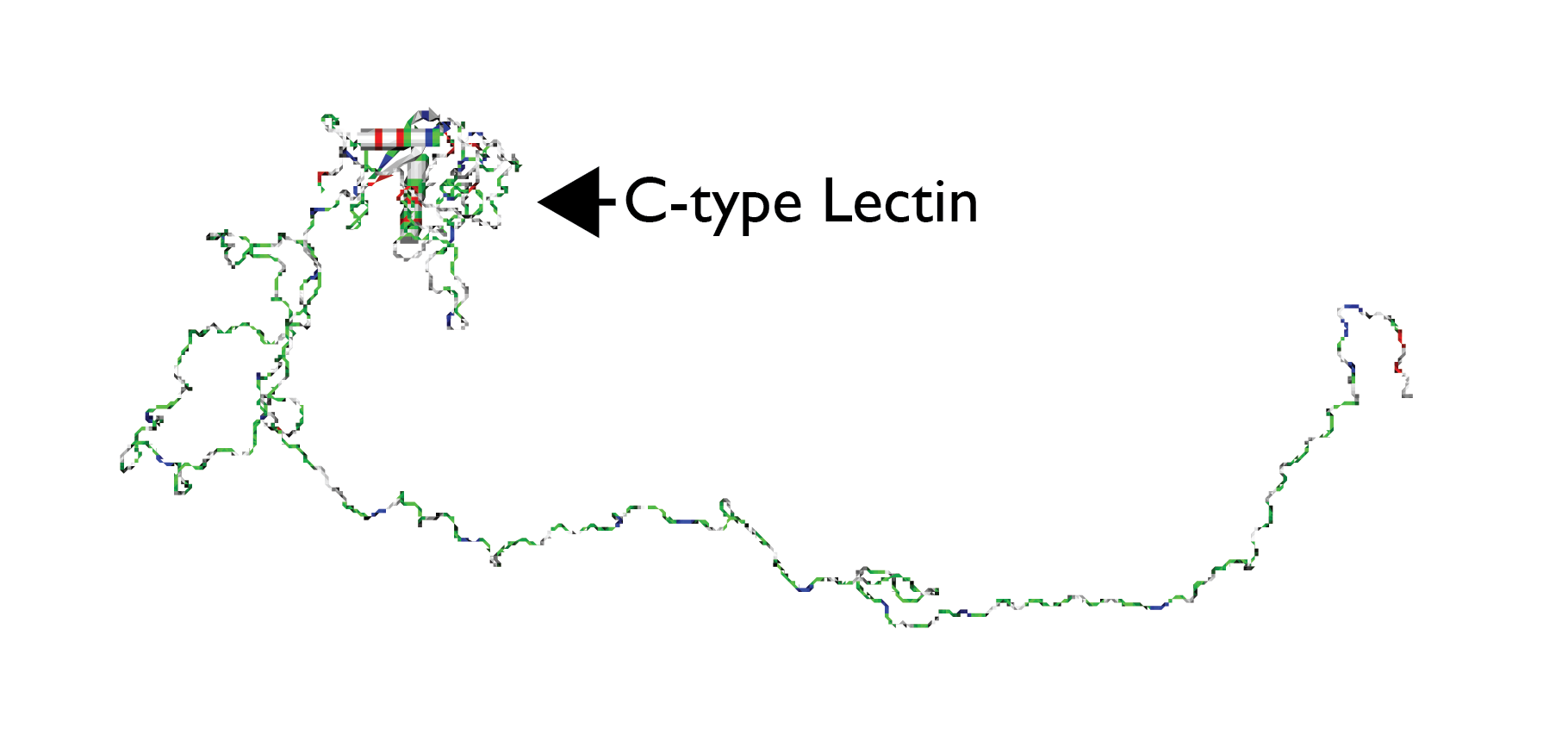
4. Summary
From the foregoing, we can observe that glycosylation provides an additional degree of control over extracellular protein function by either accentuating or attenuating the intrinsic functionality of the polypeptide sequence. In a sense, the cell can have the “last word” as to the degree of participation within the biomineralization process. In some cases (e.g., AP24), the oligosaccharides stabilize the conformation of the glycoprotein, which is a known trait of N-linked oligosaccharides [12][13][14][15][16][12-16]. The author proposes that glycosylation can serve several purposes vis a vis the biomineralization process: 1) “tweak” or “tune” protein mineralization function to suit the situation or need; 2) act as a site for molecular recognition and binding with other matrix proteins; 3) conformationally stabilize a protein, thereby enhancing functionality; 4) create additional anionic sites for ionic [e.g., Ca(II)], mineral, or water interactions; 5) invoke cell activation or deactivation via binding to outer membrane receptor proteins. Clearly, there may be other benefits that arise from glycosylation, and thus this process represents a powerful method that cells can exploit to create skeletal elements under ambient or extreme conditions [1][2][1,2}.
The biomineralization field is still in its infancy with regard to understanding the role that glycoproteins and their associated oligosaccharides play in the skeletal formation process. It is hoped that additional studies will provide more details about theser modified proteins.
5. References
1 Studart, A.R. Towards high-performance bioinspired composites. Adv. Materials 2012, 24, 5024-5044.
2 Lowenstam, H.A.; Weiner, S. On Biomineralization. Oxford University Press, New York, NY, USA 1989, pp 1-134, ISBN 0-19-504977-2.
3 Mann, S. Biomineralization. Principles and Concepts in Bioinorganic Materials Chemistry. Oxford University Press, New York, NY, USA, 2001, pp. 6-9, 24-108.
4 Wallace, A.F.; Hedges, L.O.; Fernandez-Martinez, A.; Raiteri, P.; Gale, J.D.; Waychunas, G.A.; Whitelam, S.; Banfield, J.F.; De Yoreo, J.J. Microscopic evidence for liquid-liquid separation in supersaturated calcium carbonate solutions. Science 2013, 341, 885-889.
5 De Yoreo, J.J., Gilbert, P.U.P.A., Sommerdijk, N.A.J.M, Penn, R.L., Whitelam, S., Joester, D., Zhang,H., Rimer, J.D., Navrotsky, A., Banfield, J.F., Wallace, A.F., Michel, F.M., Meldrum, F.C., Cölfen, H., Dove, P.M. Crystallization by particle attachment in synthetic, biogenic, and geologic environments. Science 2015, 349, 498-508.
6 Perovic, I., Chang, E.P., Verch, A., Rao, A., Cölfen, H., Kroeger, R., Evans, J.S. An oligomeric C-RING nacre protein influences pre-nucleation events and organizes mineral nanoparticles. Biochemistry 2014, 53, 7259-7268.
7 Liu, X., Li, J., Xiang, L., Sun, J., Zheng, G., Zhang, G., Wang, H., Xie, L., Zhang, R. The role of matrix proteins in the control of nacreous layer deposition during pearl formation. Proc. R. Soc. B 2012, 279, 1000-1007.
8 Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F., Yang, P., Zhang, L., Wang, X., Qi, H., Xiong, Z., Que, H., Xie, Y., Holland, P.W.H., Wang, X., Paps, J., Zhu, Y., Wu, F., Chen, Y., Wang, J., Peng, C., Meng, J., Yang, L., Liu, J., Wen, B., Zhang, N., Huang, Z., Zhu, Q., Feng, Y., Mount, A., Hedgecock, D., Xu, Z., Liu, Y., Domazet-Loso, T., Du., Y., Sun, X., Zhang, S., Liu, B., Cheng, P., Jiang, X., Li, J., Fan, D., Wang, W., Fu, W., Wang, T., Wang, B., Zhang, J., Peng, Z., Li, Y., Li, N., Wang, J., Chen, M., He, Y., Tan, F., Song, X., Zheng, Q., Huang, R., Yang, H., Du, X., Chen, L., Yang, M., Gaffney, P.M., Wang, S., Luo, L., She, Z., Ming, Y., Huang, W., Zhang, S., Huang, B., Zhang, Y., Qu, T., Ni, P., Miao, G., Wang, W., Zhang, S., Haung, B., Zhang, Y., Qu, T., Ni, P., Miao, G., Wang, J., Wang, Q., Steinberg, C.E.W., Wang, H., Li, N., Qian, L., Zhang, G., Li, Y., Yang, H., Liu, X., Wang, J., Yin, Y., Wang, J., The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49-54.
9 Sea urchin genome sequencing consortium et. al., The genome of the sea urchin Strongylocentrotus purpuratus. Science 2006, 314, 941-952.
10 Tirichine, L., Rastogi, A. Bowler, C. Recent progress in diatom genomics and epigenomics. Current Op. Plant Biol. 2017, 36, 46-55.
11 Uversky, V.N. Post-Translational Modification, in Brenner’s Encyclopedia of Genetics. Eds Stanley Malloy and Kelly Hughes, 2nd Ed, 2013, pp 425-430.
12 Seo, J,. Lee, K.J. Post-translational modifications and their biological functions: Proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 2004, 37, 35-44.
13 Wang, Y.C., Peterson, S.E., Loring, J.F. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Research 2014, 24, 143-160.
14 Reily, C., Stewart T.J., Renfrow, M.B. Novak, J. Glycosylation in health and disease. Nature Rev. Nephrology 2019, 15, 346-366.
15 Krasnova, L., Wong, C.H. Understanding the chemistry and biology of glycosylation with glycan synthesis. Ann. Rev. Biochem. 2016, 85, 599-630
16 Dwek, R.A., Chapter 7: Glycobiology, a quantum leap in carbohydrate chemistry, in Foundations of Modern Biochemistry 1996, Vol 2, 153-202.
17 Iijima, M., Fan, D., Bromley, K., Sun, Z., Moradian-Oldak, J. Tooth enamel proteins enamelin and amelogenin cooperate to regulate the growth morphology of octacalcium phosphate crystals. Cryst. Growth Des. 2011, 19, 4815-4822
18 Yamakoshi, Y. Carbohydrate moieties of porcine 32 kDa enamelin. Calcif. Tiss. Int. 1996, 56, 323-330.
19 Stapane, Le Roy, N., Hincke, M.T., Gautron, J. The glycoproteins EDIL3 and MFGE8 regulate vesicle-mediated eggshell calcification in a new model for avain biomineralization. J. Biol. Chem. 2019, 294, 14256-14545.
20 Couchman, J.R., Pataki, C.A. An introduction to proteoglycans and their localization. J. Histochem. Cytochem. 2012, 60, 885-897.
21 Ozawa, H., Hoshi, K., Amizuka, N. Current concepts in bone biomineralization. J. Oral Biosci. 2008, 50, 1-14.
22 Staines, K.A., MacRae, V.E., Farquharson, C. The importance of the SIBLING family of proteins on skeletal mineralization and bone remodeling. J. Endocrinology 2012, 214, 241-255.
23 Bellahcene, A., Castronovo, V., Ogbureke, K.U.E., Fisher, L.W., Fedarko, N.S. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): Multifunctional proteins in cancer. Natl. Rev. Cancer 2008, 8, 212-226.
24 Killian, C., Croker, L., Wilt, F.H. SpSM30 gene family expression in embryonic and adult biomineralized tissues of the sea urchin, Strongylocentrotus purpuratus. Gene Expression Patterns 2010, 10, 135-139.
25 Michenfelder, M., Fu, G., Lawrence, C., Weaver, J.C., Wustman, B.A., Taranto, L., Evans, J.S., Morse, D.E. Characterization of two molluscan crystal-modulating biomineralization proteins and identification of putative mineral binding domains. Biopolymers 2003, 70, 522-533; errata 73: 522.
26 Chang, E.P., Perovic, I., Rao, A., Cölfen, H., Evans, J.S. Insect cell glycosylation and its impact on the functionality of a recombinant intracrystalline nacre protein, AP24. Biochemistry 2016, 55, 1024-1035.
27 Jain, G., Pendola, M., Rao, A., Cölfen, H., Evans, J.S. A model sea urchin spicule matrix protein self-associates to form mineral-modifying protein hydrogels. Biochemistry 2016, 55, 4410-4421.
28 Juan-Colas, J, Jung, Y.S., Johnson, S, Evans, J.S A complicated relationship: Glycosylation, Ca(II), and primary sequence affect the interactions and kinetics between two model mollusk shell intracrystalline nacre proteins. Biochemistry 2020, 59, 346-350.
29 Jain, G., Pendola, M., Koutsoumpeli, E., Johnson, S., Evans, J.S. Glycosylation fosters interactions between model sea urchin spicule matrix proteins. Implications for embryonic spiculogenesis and biomineralization. Biochemistry 2018, 57, 3032-3035.
30 Pendola, M., Jain, G., Huang, Y.-C., Gebauer, D, Evans, J.S. Secrets of the sea urchin spicule revealed: Protein cooperativity is responsible for ACC transformation, intracrystalline incorporation, and guided mineral particle assembly in biocomposite material formation. ACS Omega 2018, 3, 11823-11830.
31 Le Pabic, C., Marie, A., Marie, B., Percot, A., Bonnaud-Ponticelli, L., Lopez, P.J., Luquet, G. First proteomic analysis of the dorsal and ventral parts of the Sepia officinalis cuttlebone. J. Proteomics 2017, 150, 63-73.
32 Albeck, S., Weiner, S., Addadi, L., Polysaccharides of intracrystalline proteins modulate calcite growth in vitro. Chemistry: A European Journal 1996, 2, 278-284.
33 Arivalagan, J., Yarra, T., Marie, B., Sleight, V.A., Duvernois-Berthet, E., Clark, M.S., Marie, A., Berland, S. Insights from the shell proteome: Biomineralization to adaptation. Mol. Biol and Evol. 2017, 34, 66-77.
34 Tweedie, E.P., Coblentz, F.E., Shafer, T.H. Purification of a soluble glycoprotein from the uncalcified ecdysial cuticle of the blue crab Callinectes sapidus and its possible role in initial mineralization. J. Expt. Zool. 2004, 207, 2589-2598.
35 Kamiya, H., Jimbo, M., Yako, H., Muramoto, K. Participation of the C-type hemolymph lectin in mineralization of the acorn barnacle Megabalanus rosa. Marine Biol. 2002, 140, 1235-1240.
36 Kroger, N., Bergsdorf, C., Sumper, M. A new calcium binding glycoprotein family constitutes a major diatom cell wall component. EMBO J 1994, 13, 4676-4683.
37 Nemoto, M., Ren, D., Herrera, S., Pan, S., Tamura, T., Inagaki, K. Kisailus, D. Integrative transcriptomic and proteomic analyses of a molecular mechanism of radular teeth biomineralization in Cryptochiton Stelleri. Scientific Reports 2019, 9, DOI:10.1.038/s41598-018-37839-2
