Polygenic diseases, which are genetic disorders caused by the combined action of multiple genes, pose unique and significant challenges for the diagnosis and management of affected patients. A major goal of cardiovascular medicine has been to understand how genetic variation leads to the clinical heterogeneity seen in polygenic cardiovascular diseases (CVDs). Recent advances and emerging technologies in artificial intelligence (AI), coupled with the ever-increasing availability of next generation sequencing (NGS) technologies, now provide researchers with unprecedented possibilities for dynamic and complex biological genomic analyses. Combining these technologies may lead to a deeper understanding of heterogeneous polygenic CVDs, better prognostic guidance, and, ultimately, greater personalized medicine. Advances will likely be achieved through increasingly frequent and robust genomic characterization of patients, as well the integration of genomic data with other clinical data, such as cardiac imaging, coronary angiography, and clinical biomarkers.
- genomics
- AI
- genetics
- deep learning
- cardiovascular disease
- cardiology
- machine learning
- artificial intelligence
1. Introduction
2. Genetic Testing Gap in Cardiovascular Diseases
The majority of CVDs and cardiovascular risk factors have a significant genetic component, which is most commonly polygenic in origin [1][2]. Current clinical practice utilizes a patient’s medical history, family history, physical examination, cardiac biomarkers, and various modalities of cardiac imaging to establish diagnoses and to stratify risks. Despite rapid advances and availability of genetic testing panels, clinicians seldom utilize genetic testing as part of their initial patient assessments beyond cases with a known family history of genetic, inherited CVDs (e.g., HCM, arrhythmogenic right ventricular cardiomyopathy (ARVC), long QT syndrome (LQTS), or catecholaminergic polymorphic ventricular tachycardia (CPVT)). This lack of routine testing as part of care pathway creates a “diagnostic gap” (i.e., a delay in time from disease manifestation to establishing a definitive diagnosis) that can lead to inappropriate or ineffective treatment in patients suffering from inherited CVDs. Despite its demonstrated clinical relevance, current guidelines only recommend genomic testing for a small number of cardiac conditions (e.g., HCM, familial hypercholesterolemia), limited by the relatively few genetic tests that are currently available and the lack of strong studies in cardiovascular genetics [10][11]. For example, Brugada syndrome has a large number of potentially pathogenic genetic variants (e.g., CACNA1C, GPD1L, HEY2, PKP2, RANGRF, SCN10A, SCN1B, SCN2B, SCN3B, SLMAP, and TRPM4) but current guidelines continue to recommend a comprehensive genetic analysis for only Brugada syndrome caused by the SCN5A genetic variant [12][13]. With advancements in genetic testing technologies, preemptive genetic testing for various cardiomyopathies may be useful in the presence of an asymptomatic type 1 Brugada ECG pattern, family history of dilated cardiomyopathy, or the development of spontaneous coronary artery dissection (SCAD).3. Next Generation Sequencing (NGS) in the Modern Clinic
Genomics is becoming nearly ubiquitous in biomedical research [14]. Large-scale sequencing efforts have revolutionized our understanding of the complex genetic interrelationships involved in the pathogenesis of most cardiovascular conditions [15]. The tremendous advancements in genomic research are largely driven by the advent of NGS, which has led to the discovery of novel associations and the ability to more easily assess genetic heterogeneity across patients. Several categories of NGS include: (1) whole genome sequencing (WGS); (2) whole exome sequencing (WES), where the sequencing is concentrated over the protein-coding regions of the genome (~2% of the genome); and (3) gene panels, where very deep coverage (>100× coverage) is generated for a select number of genes. Both WGS and WES allow for the accurate identification of single-nucleotide variants (SNVs), large copy number variations (CNVs), small insertion deletions (InDels), and information on variant frequencies in different populations [16]. Because WGS examines the noncoding regions of the genome, it offers a more comprehensive appraisal of both small and large genomic risk variants for CVDs. However, WGS is more costly and time-consuming than WES, and may be limited by lower depth [17][18]. Conversely, the results of WES, while more limited in scope, are typically viewed as more straightforward to interpret and historically have been a useful method to identify variants causing Mendelian disease. Panel-based NGS relies on high sequencing depth of previously determined important genetic loci, making this kind of testing more resource-efficient. However, the narrow focus of this type of assay results in decreased power to detect novel associations and is often less effective for assessing other types of genetic alterations, such as structural variants. Although NGS is now widely used due to its speed, robustness, and cost-effectiveness, orthogonal confirmation with the traditional Sanger sequencing method is sometimes still required for validation prior to clinical use [19][20][21]. Nonetheless, the implementation of AI to NGS and genomics has already been shown to accurately predict the consequences of genetic risk factors in CVDs [22][23], show the noncoding-variant effects in CVDs [24][25], find patients with cardiac amyloidosis [26][27], and initiate specific therapies from tumor sequencing [28] by integrating with electronic health records (EHRs) in several academic and medical institutions. Additionally, there are several direct-to-consumer genomics companies that use AI along with WGS and WES; however, to date, these applications have been limited by a lack of transparency in the algorithms they utilize due to their proprietary nature and commercial competition, as well as a lack of a consistent validation cohort, genomic guided clinical trials, and high-quality phenotype data that are consistently encoded and managed. Although some direct-to-consumer companies have collaborated with academic institutions and published their methodologies, evidence for their clinical relevance remains scarce.4. Introduction of AI to Clinical Cardiovascular Genetics
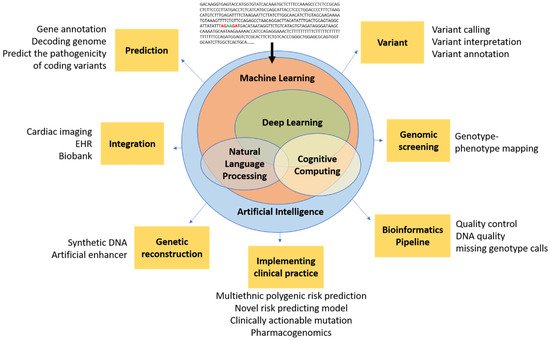
AI encompasses a broad range of applications for automated reasoning and inference, and is starting to have a major impact on clinical assessment and diagnosis. For example, in both United States of America (US) and United Kingdom (UK) datasets, AI outperformed human radiologists in screening mammography (greater than the AUC-ROC for the average radiologist by an absolute margin of 11.5%) and significantly reduced false positives and false negatives [29]. The most widely used groups of methods for pattern recognition in genomics include machine learning (ML) and deep learning (DL). Other AI approaches, for example natural language processing (NLP) and cognitive computing, are also starting to play a role in cardiovascular clinical care to enable more natural interactions between clinicians and computational systems [30][31][32]. Notably, the Food and Drug Administration (FDA) has been rapidly approving AI/ML-based medical devices and algorithms. Therefore, it is crucial for medical professionals to understand how best to utilize them. In a recent study using a web-based search for announcements of FDA approvals of AI/ML-based medical devices and algorithms, of the 64 found, 30 (46.9%), 16 (25.0%), and 10 (15.6%) were developed for the fields of radiology, cardiology, and internal medicine/general practice, respectively [33]. These AI approaches fundamentally work to train programs to recognize relationships within data. Table 1 provides examples of variant calling, reporting, and interpretation AI. Figure 1 demonstrates the potential of AI in cardiovascular genetics.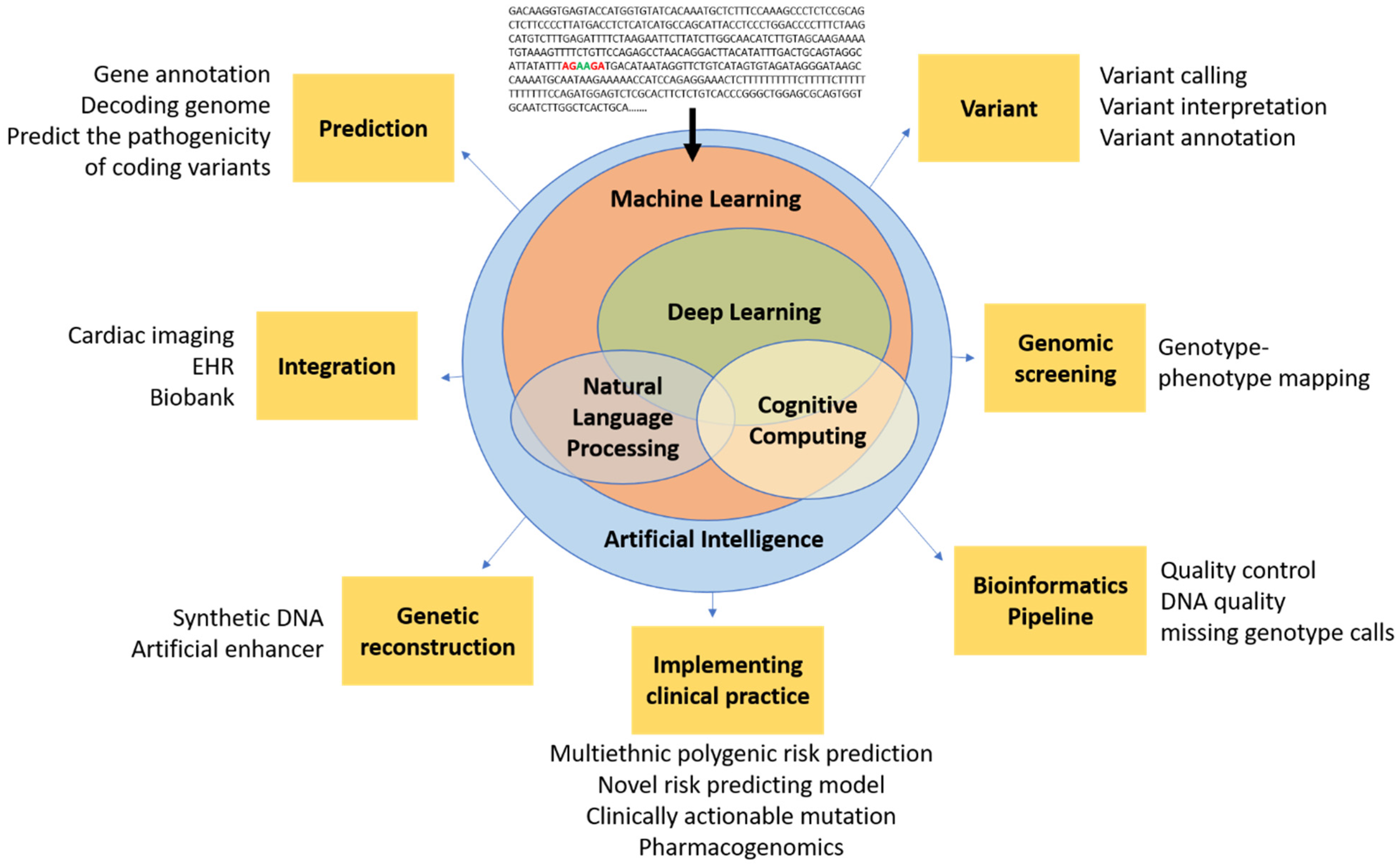
| Name | Algorithms | Example Function |
|---|---|---|
| DeepVariant [34] | Deep convolutional neural network (CNN) | Variant calling from short-read sequencing by reconstructing DNA alignments as an image |
| Clairvoyante [35] | A multi-task convolutional deep neural network | (1) Variant calling in single molecule sequencing (2) Predicts variant types (SNP or indel), zygosity, and alleles at the same time |
| Skyhawk [36] | Neural network | Mimics the process of expert review for clinically significant genomics variants identification |
| DeepBind [37] | Deep CNN | Predicts the binding sites of DNA-binding proteins and RBPs |
| iDeep [38] | Deep belief networks (DBN) and CNN | Cross-domain features and sequence information |
| DeepSEA [39] | Deep CNN | Predicts functional consequences of noncoding variants |
| DeepNano [40] | Recurrent neural networks (RNN) | Base calling in MinION nanopore reads |
| SpliceAI [41] | Deep neural network (DNN) | (1) Predicts splice junctions from an arbitrary pre-mRNA transcript sequence (2) Predicts noncoding genetic variants that cause cryptic splicing |
| DeepGestalt [42] | DNN | Distinguishes more than 200 rare diseases based on patient face images, which could also separate different genetic subtypes (e.g., Noonan syndrome) |
| DeepPVP [43] | DNN | Variant prioritization by integrating patients’ phenotype information |
| DeepSVR [44] | Deep learning and random forest models | Predicts somatic variants confirmed by orthogonal validation sequencing data |
| DeepGene [45] | DNN | Extracts the high-level features between combinatorial somatic point mutations and cancer types. Classify cancer type |
| Deep AE [46] | Autoencoder | gene expression data |
| DeepMethyl [47] | Predicts methylation states of DNA CpG dinucleotides | |
| BioVec [48] | Feature representation | |
| DeepMotif [49] | Deep convolutional/highway MLP framework | Sequential data about gene regulation |
| DeepChrome [50] | Deep CNN | Sequential data about gene regulation Classifies gene expression using histone modification data as input. |
| Chiron [51] | Deep learning model | Translates the raw signal to DNA sequence |
| Variational Autoencoders [52] | Autoencoder | Predicts drug response |
| GARFIELD-NGS [53] | Deep CNN | Dissects false and true variants in exome sequencing |
| DeepGS [54] | Deep CNN | Predicts phenotypes from genotypes |
| DANN [55] | DNN | Predicts deleterious annotation or pathogenicity of genetic variants |
| DanQ [56] | Hybrid model Deep RNN and CNN | Quantifies the function of non-coding DNA |
| ProLanGO [57] | RNN | Protein function prediction |
| BCC-NER [58] | NLP | Bidirectional and contextual clues named entity tagger for gene/protein mention recognition |
| BioNLP [59] | NLP | Gene regulation network |
| SpaCy [60] | NLP | Tagging, parsing, and entity recognition |
5. Current Limitations in Genomics and Potential Solutions with AI
5.1. Lack of Clinical and Technical Guidelines for Cardiovascular Genetics
Currently in clinical cardiovascular genetics, the guidelines do not specify which genes should be tested or how to validate the results. For example, the 2019 HRS Expert Consensus Statement on Evaluation, Risk Stratification, and Management of Arrhythmogenic Cardiomyopathy did not define how genetic testing should be validated or carried out in ARVC and other arrhythmogenic cardiomyopathies [61]. Similarly, the 2020 and 2021 scientific statements from the AHA on Genetic Testing for Heritable Cardiovascular Diseases in adult and pediatric patients did not specify how genetic testing should be validated or carried out in heritable cardiovascular diseases [7][8]. At a more rudimentary level, the Clinical Laboratory Improvement Amendment (CLIA) and the College of American Pathologists (CAP) have left many inconsistencies and regulatory gaps in their guidance for wet and dry labs [62], resulting in heterogeneous variant reporting. Moreover, CAP/CLIA regulations only require that validation is performed in the production environment, which may lead to unexpected errors in the production phase. Bioinformatics pipelines should be validated and tested for how precisely and sensitively variants are called in wet labs. Technical variability in the QC process, such as consistency of sequencing [63], QC standardization [64], and DNA quality [65][66], has been highly problematic; however, with current technologies, the accuracy of SNV is generally very robust (particularly if 30x or greater sequencing coverage is available). Another major barrier to current cardiovascular genetic research is the lack of professional recommendations for the clinical integration of genomics. Several clinical research projects using different genomics databases (e.g., UK Biobank [67], MESA [68], and ARIC [69]) have demonstrated accurate ML model discrimination and calibration (e.g., Brier score) for CVD risk prediction using genetics, but there are as yet no specific guidelines for genetic testing in clinical practice or regulatory guidance for direct-to-consumer products.5.2. Variant Calling, Reporting, and Interpretation
Variant calling is used to identify the differences between an individual genome and a reference genome. Despite CLIA approval, there are no guidelines for approval of informatics pipelines for variant calling. There are several variant-related tasks (e.g., read alignment, variant calling, reporting, and interpretation) currently used in genomics screening, the identification of probands, and cascade testing in CVD where AI could be applied. The discrepancies in variant calling between labs, largely because of the lack of clear guidelines, are magnified when undertaking the task of distinguishing true genetic variants from spurious differences introduced by sequencing errors, alignments errors, and other technical artifacts. Other limitations of variant calling include a lack of consensus between variant calling pipelines when analyzing the same data [70], variable accuracies of variant calling algorithms when using different AI technologies, and comparison sequencing of only a limited gene panel. Importantly, AI-driven software, such as DeepVariant, Clairvoyante [35], and Skyhawk [36], have already been used to automatically recognize and prioritize variants with substantially improved accuracy when compared to more traditional statistical models. For example, Google’s DeepVariant uses image recognition techniques and pre-trained models (e.g., inception-v3, variants of CNN model [71]) to pre-process inputs, make inferences, call variants, and then output variant calling format (VCF) files with the variant information. This represents a potential AI solution to the current inconsistencies in variant calling.5.3. Combining Genomics with Other Clinical Data Types
Cardiovascular genetics is challenging because both the clinical variables associated with CVDs and the genomics data are heterogeneous and often involve complex interactions between a patient’s genetics and environmental factors. This challenge is largely why applying AI to these multiple types of data is a very promising research direction, and may be especially useful in classifying genome-phenome relationships in CVD using EHRs [72]. For example, combining genomic data describing different septal morphologies of HCM [73][74] with clinical information from echocardiography and angiography could help personalize therapy for individual patients (e.g., deciding if a particular HCM patient needs an ICD). Echo-guided genetic testing or genetic-guided PCI [75] and DAPT duration (e.g., high- vs. low-risk bleeding loci) would also be useful applications of this technology. Another potential application worth researching is the diagnosis of diastolic dysfunction using a combination of echo parameters (e.g., LAVI, E/A ratio, annular e’ velocity, and peak TR velocity) and genetic predispositions since normal diastolic function changes with age [76][77][78]. Precision statin therapy is another potential application for the integration of multiple data types by AI. For instance, in a young female without traditional atherosclerotic risk factors, a combination of genetic testing (e.g., Lp (a), apo C genes) and cardiac imaging (e.g., coronary CT) may reveal a clinical need for preventative statin therapy, which would otherwise never be considered.5.4. Lack of Population Specific Analysis Tools
Across all fields of medicine and research, population-specific analysis tools and databases that can detect population-specific risk factors are urgently needed. Unfortunately, in most cases, including in CV research, significant disparities in research for different ethnicities remain. The pooled cohort equations (PCE) is the cornerstone for atherosclerotic cardiovascular disease (ASCVD) risk stratification and statin treatment decisions [11]. However, the PCE computation mainly focuses on the Caucasian population and overestimates ASCVD risk in Asian and Hispanic populations. Although PCE computations exclude genetic components, the ethnicity disparity is not limited to cardiovascular genetic research [79]. While genomic research in Asian ancestry and African ancestry has increased in recent times [80][81], more than 90% of genomic research has been conducted in patients of mainly European ancestry [82][83]. Furthermore, while most GWAS attempts can control bias of population stratification, fully correcting for population stratification can be challenging and the lack of ethnic diversity included can affect the analysis of gene–environment interactions [84]. Therefore, a major challenge for applying AI more widely is the lack of publicly available non-European genetic databases. In addition, PRS is an emerging technique for assigning genetic risk to individual outcomes that outperforms traditional risk scores [85], but the performance of translating PRS from European ancestry to different ethnicities is largely unknown and not validated [86]. The AI technique of transfer learning could potentially be used to bridge this gap.6. Current Limitations in AI Cardiovascular Genetics
Despite steadfast advances, implementing AI in cardiovascular genomics still faces several challenges, including generalizability of results, the required construction of large genomic datasets, and limited computing power. Ultimately, the largest barrier remains the ability of clinicians to implement findings from AI studies. The first challenge that plagues AI is overfitting an algorithm to a dataset that may adversely affect the generalizability of the results. Generalizability can be partially assessed by evaluating the overfitting of a new dataset. For instance, the results of applying DL models to diabetic retinopathy could not be replicated in different datasets [87][88], and AI methods lack validation data when applied to disease-associated non-coding variants [89][90]. Despite the promise of various AI methods, genomic datasets themselves have built-in limitations: the costs incurred remains a large barrier to performing thorough studies; heterogeneous genetic conditions, such as dilated cardiomyopathy, lack known outputs; and the rarity of specific conditions results in unbalanced case-control studies. These are important limitations when considering the construction of a genomics dataset. Currently, there is not a consensus or indication for genetic testing across several entities within CVD. For patients who undergo genetic testing, the sample can undergo a variety of sequencing techniques that differ between vendors, affecting the quality of the resulting data and confounding interpretation. An equally important barrier to integrating AI study results into clinical practice is the fact that physicians currently lack the necessary access as well as education and training to interpret results from AI studies on genomic data [91][92]. To facilitate clinical adoption, AI can fill the gap in knowledge in clinical practice with automated analysis to detect clinically actionable mutations. However, there is a figurative territorial embargo which limits medical genetics to trained specialists because of the complexity of handling genomic data, rather than a democratization and availability of this technology to all clinicians and patients. Emerging technology, such as homomorphic encryption or blockchains, which can provide an immediate and transparent exchange of encrypted data simultaneously to multiple parties, may be able to fill this gap by at least ensuring data security in handling genomic data. However, there is no process for lifelong interrogation of such data, nor is there specialty infrastructure or funding processes capable of handling that. Most importantly, the main challenge is “trust” in data stewardship. AI has the promise to do automated analyses, but there is no agreement over the format, interpretation, reliability, or reproducibility of the results. Finally, the quality of genomic data between direct-to-consumer companies and clinical or academic institutions may affect the availability and accuracy of “raw data” for AI to analyze. Genotyping data from direct-to-consumer companies, even those that are CLIA certified, contain errors and potentially high false-positive rates (up to 40%) [93]. For example, there is inconsistent labelling of COL3A1 and COL5A1 mutations (known to be associated with Ehlers–Danlos syndrome and SCAD) between laboratories [93]. Therefore, standard measures for correlating and combining data from direct-to-consumer and data from clinical or academic institutions are urgently needed. Beyond the technical issues of how variants are reported, there are also substantial privacy concerns involved when sharing genetic data with a direct-to-consumer company. As a minimum, advanced encryption is certainly required to maintain patient privacy.References
- Bertolini, S.; Pisciotta, L.; Di Scala, L.; Langheim, S.; Bellocchio, A.; Masturzo, P.; Cantafora, A.; Martini, S.; Averna, M.; Pes, G.M.; et al. Genetic polymorphisms affecting the phenotypic expression of familial hypercholesterolemia. Atherosclerosis 2004, 174, 57–65.
- Krittanawong, C.; Khawaja, M.; Rosenson, R.S.; Amos, C.I.; Nambi, V.; Lavie, C.J.; Virani, S.S. Association of PCSK9 Variants with the Risk of Atherosclerotic Cardiovascular Disease and Variable Responses to PCSK9 Inhibitor Therapy. Curr. Probl. Cardiol. 2021, 101043.
- Campuzano, O.; Beltrán-Álvarez, P.; Iglesias, A.; Scornik, F.; Pérez, G.; Brugada, R. Genetics and cardiac channelopathies. Genet. Med. 2010, 12, 260–267.
- Bleumink, G.S.; Schut, A.F.; Sturkenboom, M.C.; Deckers, J.W.; van Duijn, C.M.; Stricker, B.H. Genetic polymorphisms and heart failure. Genet. Med. 2004, 6, 465–474.
- Vecoli, C.; Borghini, A.; Turchi, S.; Mercuri, A.; Andreassi, M.G. Genetic polymorphisms of miRNA machinery genes in bicuspid aortic valve and associated aortopathy. Pers. Med. 2021, 18, 21–29.
- Girdauskas, E.; Geist, L.; Disha, K.; Kazakbaev, I.; Groß, T.; Schulz, S.; Ungelenk, M.; Kuntze, T.; Reichenspurner, H.; Kurth, I. Genetic abnormalities in bicuspid aortic valve root phenotype: Preliminary results†. Eur. J. Cardio-Thorac. Surg. 2017, 52, 156–162.
- Musunuru, K.; Hershberger, R.E.; Day, S.M.; Klinedinst, N.J.; Landstrom, A.P.; Parikh, V.N.; Prakash, S.; Semsarian, C.; Sturm, A.C.; American Heart Association Council on Genomic and Precision Medicine; et al. Genetic Testing for Inherited Cardiovascular Diseases: A Scientific Statement From the American Heart Association. Circ. Genom. Precis. Med. 2020, 13, e000067.
- Landstrom, A.P.; Kim, J.J.; Gelb, B.D.; Helm, B.M.; Kannankeril, P.J.; Semsarian, C.; Sturm, A.C.; Tristani-Firouzi, M.; Ware, S.M.; on behalf of the American Heart Association Council on Genomic and Precision Medicine; et al. Genetic Testing for Heritable Cardiovascular Diseases in Pediatric Patients: A Scientific Statement From the American Heart Association. Circ. Genom. Precis. Med. 2021, 14, e000086.
- Krittanawong, C.; Zhang, H.; Wang, Z.; Aydar, M.; Kitai, T. Artificial Intelligence in Precision Cardiovascular Medicine. J. Am. Coll. Cardiol. 2017, 69, 2657–2664.
- Ommen, S.R.; Mital, S.; Burke, M.A.; Day, S.M.; Deswal, A.; Elliott, P.; Evanovich, L.L.; Hung, J.; Joglar, J.A.; Kantor, P.; et al. 2020 AHA/ACC Guideline for the Diagnosis and Treatment of Patients With Hypertrophic Cardiomyopathy. Circulation 2020, 142, e558–e631.
- Grundy, S.M.; Stone, N.; Bailey, A.L.; Beam, C.; Birtcher, K.K.; Blumenthal, R.S.; Braun, L.T.; De Ferranti, S.; Faiella-Tommasino, J.; Forman, D.E.; et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2019, 73, 3168–3209.
- Brugada, J.; Campuzano, O.; Arbelo, E.; Sarquella-Brugada, G.; Brugada, R. Present Status of Brugada Syndrome. J. Am. Coll. Cardiol. 2018, 72, 1046–1059.
- Al-Khatib, S.M.; Stevenson, W.G.; Ackerman, M.J.; Bryant, W.J.; Callans, D.J.; Curtis, A.B.; Deal, B.J.; Dickfeld, T.; Field, M.E.; Fonarow, G.C.; et al. 2017 AHA/ACC/HRS Guideline for Management of Patients With Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death. Circulation 2018, 138, e272–e391.
- McKusick, V.A.; Ruddle, F.H. Toward a complete map of the human genome. Genomics 1987, 1, 103–106.
- Novelli, G.; Predazzi, I.M.; Mango, R.; Romeo, F.; Mehta, J.L. Role of genomics in cardiovascular medicine. World J. Cardiol. 2010, 2, 428–436.
- Tang, J.; Liu, R.; Zhang, Y.-L.; Liu, M.-Z.; Hu, Y.-F.; Shao, M.-J.; Zhu, L.-J.; Xin, H.-W.; Feng, G.-W.; Shang, W.-J.; et al. Application of Machine-Learning Models to Predict Tacrolimus Stable Dose in Renal Transplant Recipients. Sci. Rep. 2017, 7, 42192.
- Belkadi, A.; Bolze, A.; Itan, Y.; Cobat, A.; Vincent, Q.B.; Antipenko, A.; Shang, L.; Boisson, B.; Casanova, J.-L.; Abel, L. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc. Natl. Acad. Sci. USA 2015, 112, 5473–5478.
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169, 1177–1186.
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467.
- Lincoln, S.E.; Truty, R.; Lin, C.-F.; Zook, J.M.; Paul, J.; Ramey, V.H.; Salit, M.; Rehm, H.L.; Nussbaum, R.L.; Lebo, M.S. A Rigorous Interlaboratory Examination of the Need to Confirm Next-Generation Sequencing–Detected Variants with an Orthogonal Method in Clinical Genetic Testing. J. Mol. Diagn. 2019, 21, 318–329.
- Hume, S.; Nelson, T.N.; Speevak, M.; McCready, E.; Agatep, R.; Feilotter, H.; Parboosingh, J.; Stavropoulos, D.J.; Taylor, S.; Stockley, T.L. CCMG practice guideline: Laboratory guidelines for next-generation sequencing. J. Med. Genet. 2019, 56, 792–800.
- Aung, N.; Vargas, J.D.; Yang, C.; Cabrera, C.P.; Warren, H.R.; Fung, K.; Tzanis, E.; Barnes, M.R.; Rotter, J.I.; Taylor, K.D.; et al. Genome-Wide Analysis of Left Ventricular Image-Derived Phenotypes Identifies Fourteen Loci Associated With Cardiac Morphogenesis and Heart Failure Development. Circulation 2019, 140, 1318–1330.
- Amarbayasgalan, T.; Park, K.H.; Lee, J.Y.; Ryu, K.H. Reconstruction error based deep neural networks for coronary heart disease risk prediction. PLoS ONE 2019, 14, e0225991.
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning–based sequence model. Nat. Methods 2015, 12, 931–934.
- Jaganathan, K.; Panagiotopoulou, S.K.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24.
- Rossi, A.; Voigtlaender, M.; Janjetovic, S.; Thiele, B.; Alawi, M.; März, M.; Brandt, A.; Hansen, T.; Radloff, J.; Schön, G.; et al. Mutational landscape reflects the biological continuum of plasma cell dyscrasias. Blood Cancer J. 2017, 7, e537.
- Kufova, Z.C.; Sevcikova, T.; Januska, J.; Vojta, P.; Boday, A.; Vanickova, P.; Filipova, J.; Growkova, K.; Jelinek, T.; Hajduch, M.; et al. Newly designed 11-gene panel reveals first case of hereditary amyloidosis captured by massive parallel sequencing. J. Clin. Pathol. 2018, 71, 687–694.
- Caravagna, G.; Giarratano, Y.; Ramazzotti, D.; Tomlinson, I.; Graham, T.A.; Sanguinetti, G.; Sottoriva, A. Detecting repeated cancer evolution from multi-region tumor sequencing data. Nat. Methods 2018, 15, 707–714.
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94.
- Krittanawong, C.; Johnson, K.W.; Hershman, S.G.; Tang, W. Big data, artificial intelligence, and cardiovascular precision medicine. Expert Rev. Precis. Med. Drug Dev. 2018, 3, 305–317.
- Johnson, K.; Shameer, K.; Glicksberg, B.; Readhead, B.; Sengupta, P.P.; Björkegren, J.L.; Kovacic, J.C.; Dudley, J.T. Enabling Precision Cardiology Through Multiscale Biology and Systems Medicine. JACC Basic Transl. Sci. 2017, 2, 311–327.
- Johnson, K.; Soto, J.T.; Glicksberg, B.; Shameer, K.; Miotto, R.; Ali, M.; Ashley, E.; Dudley, J.T. Artificial Intelligence in Cardiology. J. Am. Coll. Cardiol. 2018, 71, 2668–2679.
- Benjamens, S.; Dhunnoo, P.; Meskó, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: An online database. NPJ Digit. Med. 2020, 3, 118.
- Poplin, R.; Chang, P.-C.; Alexander, D.; Schwartz, S.; Colthurst, T.; Ku, A.; Newburger, D.; Dijamco, J.; Nguyen, N.; Afshar, P.T.; et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018, 36, 983–987.
- Luo, R.; Sedlazeck, F.J.; Lam, T.-W.; Schatz, M.C. A multi-task convolutional deep neural network for variant calling in single molecule sequencing. Nat. Commun. 2019, 10, 998.
- Luo, R.; Lam, T.-W.; Schatz, M.C. Skyhawk: An Artificial Neural Network-based discriminator for reviewing clinically significant genomic variants. bioRxiv 2019, 13, 311985.
- Hassanzadeh, H.R.; Wang, M.D. DeeperBind: Enhancing prediction of sequence specificities of DNA binding proteins. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shen Zhen, China, 15–18 December 2016; pp. 178–183.
- Pan, X.; Shen, H.-B. RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach. BMC Bioinform. 2017, 18, 136.
- DeepSea. Available online: https://hb.flatironinstitute.org/deepsea/ (accessed on 8 February 2022).
- Boža, V.; Brejova, B.; Vinař, T. DeepNano: Deep recurrent neural networks for base calling in MinION nanopore reads. PLoS ONE 2017, 12, e0178751.
- SpliceAI: Predicting Splicing from Primary Sequence with Deep Learning. Available online: https://hpc.nih.gov/apps/SpliceAI.html (accessed on 8 February 2022).
- Gurovich, Y.; Hanani, Y.; Bar, O.; Nadav, G.; Fleischer, N.; Gelbman, D.; Basel-Salmon, L.; Krawitz, P.M.; Kamphausen, S.B.; Zenker, M.; et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 2019, 25, 60–64.
- PhenomeNet Variant Predictor (PVP). Available online: https://github.com/bio-ontology-research-group/phenomenet-vp (accessed on 8 February 2022).
- Ainscough, B.J.; Barnell, E.K.; Ronning, P.; Campbell, K.M.; Wagner, A.H.; Fehniger, T.A.; Dunn, G.P.; Uppaluri, R.; Govindan, R.; Rohan, T.E.; et al. A deep learning approach to automate refinement of somatic variant calling from cancer sequencing data. Nat. Genet. 2018, 50, 1735–1743.
- Yuan, Y.; Shi, Y.; Li, C.; Kim, J.; Cai, W.; Han, Z.; Feng, D.D. DeepGene: An advanced cancer type classifier based on deep learning and somatic point mutations. BMC Bioinform. 2016, 17, 243–256.
- Xie, R.; Wen, J.; Quitadamo, A.; Cheng, J.; Shi, X. A deep auto-encoder model for gene expression prediction. BMC Genom. 2017, 18, 39–49.
- Wang, Y.; Liu, T.; Xu, D.; Shi, H.; Zhang, C.; Mo, Y.-Y.; Wang, Z. Predicting DNA Methylation State of CpG Dinucleotide Using Genome Topological Features and Deep Networks. Sci. Rep. 2016, 6, 19598.
- Abrahamsson, E.; Plotkin, S.S. BioVEC: A program for Biomolecule Visualization with Ellipsoidal Coarse-graining. J. Mol. Graph. Model. 2009, 28, 140–145.
- Lanchantin, J.; Singh, R.; Wang, B.; Qi, Y. Deep Motif Dashboard: Visualizing and Understanding Genomic Sequences Using Deep Neural Net-Works. Pac. Symp. Biocomput. 2017, 22, 254–265.
- Singh, R.; Lanchantin, J.; Robins, G.; Qi, Y. DeepChrome: Deep-learning for predicting gene expression from histone modifications. Bioinformatics 2016, 32, i639–i648.
- Teng, H.; Cao, M.D.; Hall, M.B.; Duarte, T.; Wang, S.; Coin, L.J.M. Chiron: Translating nanopore raw signal directly into nucleotide sequence using deep learning. GigaScience 2018, 7, giy037.
- Way, G.P.; Greene, C.S. Extracting a biologically relevant latent space from cancer transcriptomes with variational autoencoders. Pac. Symp. Biocomput. 2018, 23, 80–91.
- Ravasio, V.; Ritelli, M.; Legati, A.; Giacopuzzi, E. GARFIELD-NGS: Genomic vARiants FIltering by dEep Learning moDels in NGS. Bioinformatics 2018, 34, 3038–3040.
- Lin, X.; Zhao, K.; Xiao, T.; Quan, Z.; Wang, Z.-J.; Yu, P.S. DeepGS: Deep Representation Learning of Graphs and Sequences for Drug-Target Binding Affinity Prediction. arXiv 2020, arXiv:2003.13902.
- Quang, D.; Chen, Y.; Xie, X. DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 2015, 31, 761–763.
- Quang, D.; Xie, X. DanQ: A hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016, 44, e107.
- Cao, R.; Freitas, C.; Chan, L.; Sun, M.; Jiang, H.; Chen, Z. ProLanGO: Protein Function Prediction Using Neural Machine Translation Based on a Recurrent Neural Network. Molecules 2017, 22, 1732.
- BCC-NER Gene/Protein Mention Tagger. Available online: http://www.biominingbu.org:8080/BCC-NER/ (accessed on 8 February 2022).
- Provoost, T.; Moens, M.-F. Semi-supervised Learning for the BioNLP Gene Regulation Network. BMC Bioinform. 2015, 16, S4.
- Ramachandran, R.; Arutchelvan, K. Named entity recognition on bio-medical literature documents using hybrid based approach. J. Ambient Intell. Humaniz. Comput. 2021, 10, 1–10.
- Towbin, J.A.; McKenna, W.J.; Abrams, D.J.; Ackerman, M.J.; Calkins, H.; Darrieux, F.C.C.; Daubert, J.P.; de Chillou, C.; DePasquale, E.C.; Desai, M.Y.; et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardio-myopathy. Heart Rhythm 2019, 16, e301–e372.
- Tavtigian, S.V.; Greenblatt, M.S.; Harrison, S.M.; Nussbaum, R.L.; Prabhu, S.A.; Boucher, K.M.; Biesecker, L.G. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 2018, 20, 1054–1060.
- Phelan, J.; O’Sullivan, D.M.; Machado, D.; Ramos, J.; Whale, A.S.; O’Grady, J.; Dheda, K.; Campino, S.; McNerney, R.; Viveiros, M.; et al. The variability and reproducibility of whole genome sequencing technology for detecting resistance to anti-tuberculous drugs. Genome Med. 2016, 8, 132.
- Traore, K.; Bull, S.; Niare, A.; Konate, S.; Thera, M.A.; Kwiatkowski, D.; Parker, M.; Doumbo, O.K. Understandings of genomic research in developing countries: A qualitative study of the views of MalariaGEN participants in Mali. BMC Med. Ethic. 2015, 16, 42.
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661.
- Clayton, D.G.; Walker, N.M.; Smyth, D.J.; Pask, R.; Cooper, J.D.; Maier, L.M.; Smink, L.J.; Lam, A.C.; Ovington, N.R.; Stevens, H.E.; et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 2005, 37, 1243–1246.
- Alaa, A.M.; Bolton, T.; Angelantonio, E.D.; Rudd, J.H.F.; van der Schaar, M. Cardiovascular disease risk prediction using automated machine learning: A prospective study of 423,604 UK Biobank participants. PLoS ONE 2019, 14, e0213653.
- Ambale-Venkatesh, B.; Yang, X.; Wu, C.O.; Liu, K.; Hundley, W.G.; McClelland, R.; Gomes, A.S.; Folsom, A.R.; Shea, S.; Guallar, E.; et al. Cardiovascular Event Prediction by Machine Learning: The Multi-Ethnic Study of Atherosclerosis. Circ. Res. 2017, 121, 1092–1101.
- Zhuang, X.; Sun, X.; Zhong, X.; Zhou, H.; Zhang, S.; Liao, X. Deep phenotyping and prediction of long-term heart failure by machine learning. J. Am. Coll. Cardiol. 2019, 73, 690.
- O’Rawe, J.; Jiang, T.; Sun, G.; Wu, Y.; Wang, W.; Hu, J.; Bodily, P.; Tian, L.; Hakonarson, H.; Johnson, W.E.; et al. Low concordance of multiple variant-calling pipelines: Practical implications for exome and genome sequencing. Genome Med. 2013, 5, 28.
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016.
- Glicksberg, B.; Johnson, K.; Dudley, J.T. The next generation of precision medicine: Observational studies, electronic health records, biobanks and continuous monitoring. Hum. Mol. Genet. 2018, 27, R56–R62.
- Solomon, S.D.; Wolff, S.; Watkins, H.; Ridker, P.M.; Come, P.; McKenna, W.J.; Seidman, C.E.; Lee, R.T. Left ventricular hypertrophy and morphology in familial hypertrophic cardiomyopathy associated with mutations of the beta-myosin heavy chain gene. J. Am. Coll. Cardiol. 1993, 22, 498–505.
- Binder, J.; Ommen, S.R.; Gersh, B.J.; Van Driest, S.L.; Tajik, A.J.; Nishimura, R.A.; Ackerman, M.J. Echocardiography-Guided Genetic Testing in Hypertrophic Cardiomyopathy: Septal Morphological Features Predict the Presence of Myofilament Mutations. Mayo Clin. Proc. 2006, 81, 459–467.
- Claassens, D.M.F.; Vos, G.J.; Bergmeijer, T.O.; Hermanides, R.S.; Hof, A.W.V.T.; Van Der Harst, P.; Barbato, E.; Morisco, C.; Gin, R.M.T.J.; Asselbergs, F.W.; et al. A Genotype-Guided Strategy for Oral P2Y12 Inhibitors in Primary PCI. N. Engl. J. Med. 2019, 381, 1621–1631.
- Bockhorst, J.; Craven, M.; Page, D.; Shavlik, J.; Glasner, J. A Bayesian network approach to operon prediction. Bioinformatics 2003, 19, 1227–1235.
- Cawley, S.L.; Pachter, L. HMM sampling and applications to gene finding and alternative splicing. Bioinformatics 2003, 19, ii36–ii41.
- Nagueh, S.F.; Smiseth, O.A.; Appleton, C.P.; Byrd, B.F.; Dokainish, H.; Edvardsen, T.; Flachskampf, F.A.; Gillebert, T.C.; Klein, A.L.; Lancellotti, P.; et al. Recommendations for the Evaluation of Left Ventricular Diastolic Function by Echocardiography: An Update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur. Hear. J. Cardiovasc. Imaging 2016, 17, 1321–1360.
- Rodriguez, F.; Chung, S.; Blum, M.R.; Coulet, A.; Basu, S.; Palaniappan, L.P. Atherosclerotic Cardiovascular Disease Risk Prediction in Disaggregated Asian and Hispanic Subgroups Using Elec-tronic Health Records. J. Am. Heart Assoc. 2019, 8, e011874.
- Popejoy, A.B.; Fullerton, S.M. Genomics is failing on diversity. Nature 2016, 538, 161–164.
- Ng, M.C.Y.; Shriner, D.; Chen, B.H.; Li, J.; Chen, W.-M.; Guo, X.; Liu, J.; Bielinski, S.J.; Yanek, L.R.; Nalls, M.A.; et al. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 2014, 10, e1004517.
- Bustamante, C.D.; Burchard, E.G.; De la Vega, F.M. Genomics for the world. Nature 2011, 475, 163–165.
- Need, A.C.; Goldstein, D.B. Next generation disparities in human genomics: Concerns and remedies. Trends Genet. 2009, 25, 489–494.
- Shi, M.; Umbach, D.M.; Weinberg, C.R. Family-based gene-by-environment interaction studies: Revelations and remedies. Epidemiology 2011, 22, 400–407.
- Inouye, M.; Abraham, G.; Nelson, C.P.; Wood, A.M.; Sweeting, M.J.; Dudbridge, F.; Lai, F.Y.; Kaptoge, S.; Brozynska, M.; Wang, T.; et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults. J. Am. Coll. Cardiol. 2018, 72, 1883.
- Martin, A.R.; Kanai, M.; Kamatani, Y.; Okada, Y.; Neale, B.M.; Daly, M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019, 51, 584–591.
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410.
- Voets, M. Deep Learning: From Data Extraction to Large-Scale Analysis; UiT Norges Arktiske Universitet: Alta, Norway, 2018.
- Schubach, M.; Re, M.; Robinson, P.N.; Valentini, G. Imbalance-Aware Machine Learning for Predicting Rare and Common Disease-Associated Non-Coding Variants. Sci. Rep. 2017, 7, 2959.
- Giral, H.; Landmesser, U.; Kratzer, A. Into the Wild: GWAS Exploration of Non-coding RNAs. Front. Cardiovasc. Med. 2018, 5, 181.
- Klitzman, R.; Chung, W.; Marder, K.; Shanmugham, A.; Chin, L.J.; Stark, M.; Leu, C.-S.; Appelbaum, P.S. Attitudes and Practices Among Internists Concerning Genetic Testing. J. Genet. Couns. 2013, 22, 90–100.
- Giardiello, F.M.; Brensinger, J.D.; Petersen, G.M.; Luce, M.C.; Hylind, L.M.; Bacon, J.A.; Booker, S.V.; Parker, R.D.; Hamilton, S.R. The use and interpretation of commercial APC gene testing for familial adenomatous polyposis. N. Engl. J. Med. 1997, 336, 823–827.
- Tandy-Connor, S.; Guiltinan, J.; Krempely, K.; LaDuca, H.; Reineke, P.; Gutierrez, S.; Gray, P.; Davis, B.T. False-positive results released by direct-to-consumer genetic tests highlight the importance of clinical confirma-tion testing for appropriate patient care. Gene. Med. 2018, 20, 1515–1521.
