Nonlinear mixed effects models have become a standard platform for analysis when data is in the form of continuous and repeated measurements of subjects from a population of interest, while temporal profiles of subjects commonly follow a nonlinear tendency. While frequentist analysis of nonlinear mixed effects models has a long history, Bayesian analysis of the models has received comparatively little attention until the late 1980s, primarily due to the time-consuming nature of Bayesian computation. Since the early 1990s, Bayesian approaches for the models began to emerge to leverage rapid developments in computing power, and have recently received significant attention due to (1) superiority to quantify the uncertainty of parameter estimation; (2) utility to incorporate prior knowledge into the models; and (3) flexibility to match exactly the increasing complexity of scientific research arising from diverse industrial and academic fields.
- Bayesian Nonlinear Mixed Effects Models
1. Introduction
2. Trends and Workflow of Bayesian Nonlinear Mixed Effects Models
2.1. Rise in the Use of Bayesian Approaches for the Nonlinear Mixed Effects Models
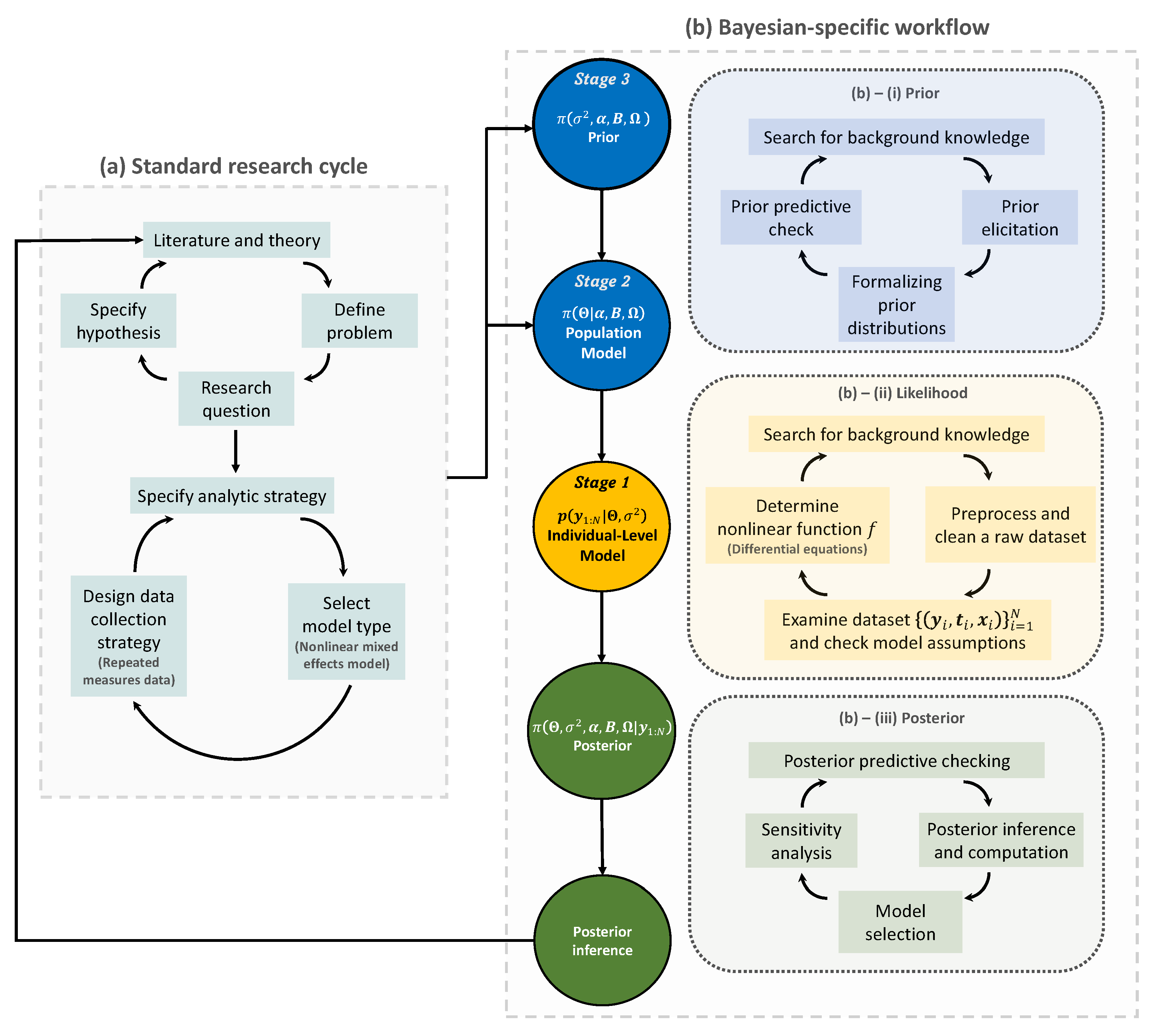
2.2. Bayesian Workflow
References
- Sterba, S.K. Fitting nonlinear latent growth curve models with individually varying time points. Struct. Equ. Model. Multidiscip. J. 2014, 21, 630–647.
- McArdle, J.J. Latent variable growth within behavior genetic models. Behav. Genet. 1986, 16, 163–200.
- Cook, N.R.; Ware, J.H. Design and analysis methods for longitudinal research. Annu. Rev. Public Health 1983, 4, 1–23.
- Mehta, P.D.; West, S.G. Putting the individual back into individual growth curves. Psychol. Methods 2000, 5, 23.
- Zeger, S.L.; Liang, K.Y. An overview of methods for the analysis of longitudinal data. Stat. Med. 1992, 11, 1825–1839.
- Diggle, P.; Diggle, P.J.; Heagerty, P.; Liang, K.Y.; Zeger, S. Analysis of Longitudinal Data; Oxford University Press: Oxford, UK, 2002.
- Demidenko, E. Mixed Models: Theory and Applications with R; John Wiley & Sons: Hoboken, NJ, USA, 2013.
- Snijders, T.A.; Bosker, R.J. Multilevel Analysis: An Introduction to Basic and Advanced Multilevel Modeling; Sage: Los Angeles, CA, USA, 2011.
- Goldstein, H. Multilevel Statistical Models; John Wiley & Sons: Hoboken, NJ, USA, 2011; Volume 922.
- Raudenbush, S.W.; Bryk, A.S. Hierarchical Linear Models: Applications and Data Analysis Methods; Sage: Thousand Oaks, CA, USA, 2002; Volume 1.
- Efron, B. The future of indirect evidence. Stat. Sci. A Rev. J. Inst. Math. Stat. 2010, 25, 145.
- Sheiner, L.B.; Rosenberg, B.; Melmon, K.L. Modelling of individual pharmacokinetics for computer-aided drug dosage. Comput. Biomed. Res. 1972, 5, 441–459.
- Lindstrom, M.J.; Bates, D.M. Nonlinear mixed effects models for repeated measures data. Biometrics 1990, 46, 673–687.
- Davidian, M.; Giltinan, D.M. Nonlinear models for repeated measurement data: An overview and update. J. Agric. Biol. Environ. Stat. 2003, 8, 387–419.
- Davidian, M.; Giltinan, D.M. Nonlinear Models for Repeated Measurement Data; Routledge: New York, NY, USA, 1995.
- Beal, S. The NONMEM System. 1980. Available online: https://iconplc.com/innovation/nonmem/ (accessed on 20 February 2022).
- Stan Development Team. RStan: The R Interface to Stan. R Package Version 2.21.3. 2021. Available online: https://mc-stan.org/rstan/ (accessed on 20 February 2022).
- Fidler, M.; Wilkins, J.J.; Hooijmaijers, R.; Post, T.M.; Schoemaker, R.; Trame, M.N.; Xiong, Y.; Wang, W. Nonlinear mixed-effects model development and simulation using nlmixr and related R open-source packages. CPT Pharmacometr. Syst. Pharmacol. 2019, 8, 621–633.
- Wang, W.; Hallow, K.; James, D. A tutorial on RxODE: Simulating differential equation pharmacometric models in R. CPT Pharmacometr. Syst. Pharmacol. 2016, 5, 3–10.
- Stegmann, G.; Jacobucci, R.; Harring, J.R.; Grimm, K.J. Nonlinear mixed-effects modeling programs in R. Struct. Equ. Model. Multidiscip. J. 2018, 25, 160–165.
- Vonesh, E.; Chinchilli, V.M. Linear and Nonlinear Models for the Analysis of Repeated Measurements; CRC Press: Boca Raton, FL, USA, 1996.
- Lee, S.Y. Structural Equation Modeling: A Bayesian Approach; John Wiley & Sons: Hoboken, NJ, USA, 2007.
- Dellaportas, P.; Smith, A.F. Bayesian inference for generalized linear and proportional hazards models via Gibbs sampling. J. R. Stat. Soc. Ser. C 1993, 42, 443–459.
- Bush, C.A.; MacEachern, S.N. A semiparametric Bayesian model for randomised block designs. Biometrika 1996, 83, 275–285.
- Zeger, S.L.; Karim, M.R. Generalized linear models with random effects; a Gibbs sampling approach. J. Am. Stat. Assoc. 1991, 86, 79–86.
- Brooks, S.P. Bayesian computation: A statistical revolution. Philos. Trans. R. Soc. Lond. Ser. A Math. Phys. Eng. Sci. 2003, 361, 2681–2697.
- Bennett, J.; Wakefield, J. A comparison of a Bayesian population method with two methods as implemented in commercially available software. J. Pharmacokinet. Biopharm. 1996, 24, 403–432.
- Wakefield, J. The Bayesian analysis of population pharmacokinetic models. J. Am. Stat. Assoc. 1996, 91, 62–75.
- Gelman, A.; Bois, F.; Jiang, J. Physiological pharmacokinetic analysis using population modeling and informative prior distributions. J. Am. Stat. Assoc. 1996, 91, 1400–1412.
- Lee, S.Y.; Lei, B.; Mallick, B. Estimation of COVID-19 spread curves integrating global data and borrowing information. PLoS ONE 2020, 15, e0236860.
- Lee, S.Y.; Mallick, B.K. Bayesian Hierarchical Modeling: Application Towards Production Results in the Eagle Ford Shale of South Texas. Sankhya B 2021, 1–43.
- Hammersley, J. Monte Carlo Methods; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013.
- Green, P.J.; Łatuszyński, K.; Pereyra, M.; Robert, C.P. Bayesian computation: A summary of the current state, and samples backwards and forwards. Stat. Comput. 2015, 25, 835–862.
- Plummer, M. JAGS: A program for analysis of Bayesian graphical models using Gibbs sampling. In Proceedings of the 3rd International Workshop on Distributed Statistical Computing, Vienna, Austria, 20–22 March 2003; Volume 124, pp. 1–10.
- Lunn, D.; Spiegelhalter, D.; Thomas, A.; Best, N. The BUGS project: Evolution, critique and future directions. Stat. Med. 2009, 28, 3049–3067.
- Chernoff, H. Large-sample theory: Parametric case. Ann. Math. Stat. 1956, 27, 1–22.
- Beal, S.L.; Sheiner, L.B.; Boeckmann, A.; Bauer, R.J. NONMEM Users Guides; NONMEM Project Group, University of California: San Francisco, CA, USA, 1992.
- Lavielle, M. Monolix User Guide Manual. 2005. Available online: https://monolix.lixoft.com/ (accessed on 20 February 2022).
- Wand, M. Fisher information for generalised linear mixed models. J. Multivar. Anal. 2007, 98, 1412–1416.
- Kang, D.; Bae, K.S.; Houk, B.E.; Savic, R.M.; Karlsson, M.O. Standard error of empirical bayes estimate in NONMEM® VI. Korean J. Physiol. Pharmacol. 2012, 16, 97–106.
- Breslow, N.E.; Clayton, D.G. Approximate inference in generalized linear mixed models. J. Am. Stat. Assoc. 1993, 88, 9–25.
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Rubin, D.B. Bayesian Data Analysis; Chapman and Hall/CRC: London, UK, 2004.
- Smid, S.C.; McNeish, D.; Miočević, M.; van de Schoot, R. Bayesian versus frequentist estimation for structural equation models in small sample contexts: A systematic review. Struct. Equ. Model. Multidiscip. J. 2020, 27, 131–161.
- Rupp, A.A.; Dey, D.K.; Zumbo, B.D. To Bayes or not to Bayes, from whether to when: Applications of Bayesian methodology to modeling. Struct. Equ. Model. 2004, 11, 424–451.
- Bonangelino, P.; Irony, T.; Liang, S.; Li, X.; Mukhi, V.; Ruan, S.; Xu, Y.; Yang, X.; Wang, C. Bayesian approaches in medical device clinical trials: A discussion with examples in the regulatory setting. J. Biopharm. Stat. 2011, 21, 938–953.
- Campbell, G. Bayesian methods in clinical trials with applications to medical devices. Commun. Stat. Appl. Methods 2017, 24, 561–581.
- Hoff, P.D. A First Course in Bayesian Statistical Methods; Springer: Berlin/Heidelberg, Germany, 2009; Volume 580.
- O’Hagan, A. Bayesian statistics: Principles and benefits. Frontis 2004, 3, 31–45.
- van de Schoot, R.; Depaoli, S.; King, R.; Kramer, B.; Märtens, K.; Tadesse, M.G.; Vannucci, M.; Gelman, A.; Veen, D.; Willemsen, J.; et al. Bayesian statistics and modelling. Nat. Rev. Methods Prim. 2021, 1, 1–26.
- Blaxter, L.; Hughes, C.; Tight, M. How to Research; McGraw-Hill Education: New York, NY, USA, 2010.
- Neuman, W.L. Understanding Research; Pearson: New York, NY, USA, 2016.
- Pinheiro, J.; Bates, D. Mixed-Effects Models in S and S-PLUS; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006.
- Gelman, A.; Simpson, D.; Betancourt, M. The prior can often only be understood in the context of the likelihood. Entropy 2017, 19, 555.
- Garthwaite, P.H.; Kadane, J.B.; O’Hagan, A. Statistical methods for eliciting probability distributions. J. Am. Stat. Assoc. 2005, 100, 680–701.
- O’Hagan, A.; Buck, C.E.; Daneshkhah, A.; Eiser, J.R.; Garthwaite, P.H.; Jenkinson, D.J.; Oakley, J.E.; Rakow, T. Uncertain Judgements: Eliciting Experts’ Probabilities; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006.
- Howard, G.S.; Maxwell, S.E.; Fleming, K.J. The proof of the pudding: An illustration of the relative strengths of null hypothesis, meta-analysis, and Bayesian analysis. Psychol. Methods 2000, 5, 315.
- Levy, R. Bayesian data-model fit assessment for structural equation modeling. Struct. Equ. Model. Multidiscip. J. 2011, 18, 663–685.
- Wang, L.; Cao, J.; Ramsay, J.O.; Burger, D.; Laporte, C.; Rockstroh, J.K. Estimating mixed-effects differential equation models. Stat. Comput. 2014, 24, 111–121.
- Botha, I.; Kohn, R.; Drovandi, C. Particle methods for stochastic differential equation mixed effects models. Bayesian Anal. 2021, 16, 575–609.
- Fucik, S.; Kufner, A. Nonlinear Differential Equations; Elsevier: Amsterdam, The Netherlands, 2014.
- Verhulst, F. Nonlinear Differential Equations and Dynamical Systems; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006.
- Cohen, S.D.; Hindmarsh, A.C.; Dubois, P.F. CVODE, a stiff/nonstiff ODE solver in C. Comput. Phys. 1996, 10, 138–143.
- Dormand, J.R.; Prince, P.J. A family of embedded Runge-Kutta formulae. J. Comput. Appl. Math. 1980, 6, 19–26.
- Margossian, C.; Gillespie, B. Torsten: A Prototype Model Library for Bayesian PKPD Modeling in Stan User Manual: Version 0.81. Available online: https://metrumresearchgroup.github.io/Torsten/ (accessed on 20 February 2022).
