Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 1 by Mohammad Imran.
Antibiotic resistance has become a threat to microbial therapies nowadays. The conventional approaches possess several limitations to combat microbial infections. Therefore, to overcome such complications, novel drug delivery systems have gained pharmaceutical scientists’ interest. Significant findings have validated the effectiveness of novel drug delivery systems such as polymeric nanoparticles, liposomes, metallic nanoparticles, dendrimers, and lipid-based nanoparticles against severe microbial infections and combating antimicrobial resistance.
- antimicrobial resistance
- nanoparticles
1. Introduction
Utilizing nanoparticulate materials to treat infectious diseases has been the subject of great interest in recent times. Infection resulting from multidrug resistance organisms (MDROs) are among the significant causes of morbidity and mortality worldwide [1]. In addition, antibiotics available for the treatment of MDROs are limited. Nevertheless, the development of new antibiotics to tackle MDROs infection requires huge economic and personnel investment and is laborious. Therefore, these critical clinical challenges emphasize the development of alternative and effective antimicrobial strategies to treat MDROs infections. Notably, owing to their unique physiochemical attributes, nanoparticles demonstrate therapeutic promise. Nanoparticles could be categorized in two ways: organic (e.g., lipid-based nanoparticle) or inorganic (e.g., nanoparticles of metals), and these have proved highly efficacious in treating various health complications [2]. Notably, organic nanoparticles have been widely reported to increase the bioavailability of drugs, enhance penetration and drug delivery, and improve antibacterial activities. The most attractive aspect of nanoparticles is the ability to incorporate different types of therapeutics, whether attached to their surface or incorporated within the structure; therefore, increasing the availability of drugs to the site of the action and better efficacy. In addition, nanomaterials can disrupt the bacterial cell membrane and target intracellular components and inhibit the proper functioning of the cellular machinery of microbes.
In 2008, the American Society of Infectious Diseases coined the term ESKAPE to categorize the deadly bacterial pathogens growing at pace and adapting to be multidrug-resistant against different antibiotics. The pathogens included Enterococcus, Staphylococcus, Klebsiella, and Pseudomonas species. The “escape” mechanism and behavior of these pathogens from the biocidal effect of contemporary marketed therapeutics causes life-threatening serious complications such as hospital-acquired infections [3]. Macrophages are responsible for eradicating foreign microbes, pathogens, and particles from the systemic circulation via a specific pathway. These phagocytic pathways could be exploited for the delivery of nanocarriers containing antibiotics for superior therapeutic effects. These pathways enable nanoparticles to be delivered directly to macrophages. It is believed that antibiotic-loaded nanocarriers can passively accumulate via the specific identification mechanism and uptake by phagocytes. Subsequently, the uptake of nanocarriers by phagocytosis leads to the release of drug payload in the infected cells, which eventually increases the permeation and exhibits superior therapeutic effects.
Moreover, biofilm-associated antimicrobial resistance is another concern. Therefore, nanocarriers have to play a role in overcoming these hurdles for better efficacy because they act as a protective coat, shielding against interactions, reducing the inactivation of drugs by biofilm and resident enzymes. In this scenario, nano-based drug delivery systems are promising for the treatment of either intracellular or biofilm-forming pathogenies infections. Diverse nanoformulations, such as liposomes and polymeric nanoparticles (NPs), have been developed and employed to deliver antibiotics to difficult-to-treat bacteria [3].
Therefore, the application of nanoparticles in antibacterial therapy to overcome MDR is an emerging approach.
2. Antibiotic Resistance Mechanism in Bacteria
The development of antibiotic resistance in pathologically lethal microorganisms is considered one of the foremost public health challenges to tackle infectious diseases worldwide. Nosocomial infections by multidrug resistance microorganisms increase the risk of life-threatening conditions to the patient in postoperative wards, burn units, and critical care units. For instance, hospitals are the major MDROs colonizing spot [4], but these are not limited to the hospital settings only. Community sites such as animal farms, biohazardous material dumping areas, freshwater environments, etc. [5,6][5][6] are a significant breeding ground for MDROs. The lack of aseptic techniques used in patient care, such as the use of one nonsterilized stethoscope or thermometer, ungloved or single-gloved hands in multiple patients in hospital wards, unethical and improper use of antibiotics in animal farms, dumping of unsterilized hospital waste in dumping sites and freshwater sources are increasing the risk of the spread of MDROs, in community environments. In this way, the horizontal transfer of resistant genes into surrounding microorganisms is also hastened. According to the nature of microorganisms, several mechanisms can be adopted to develop resistance toward specific antibiotics. In this topic, wHere will discuss some of is the mechanisms adopted by bacteria for the development of resistance against different antibiotics (Figure 1).
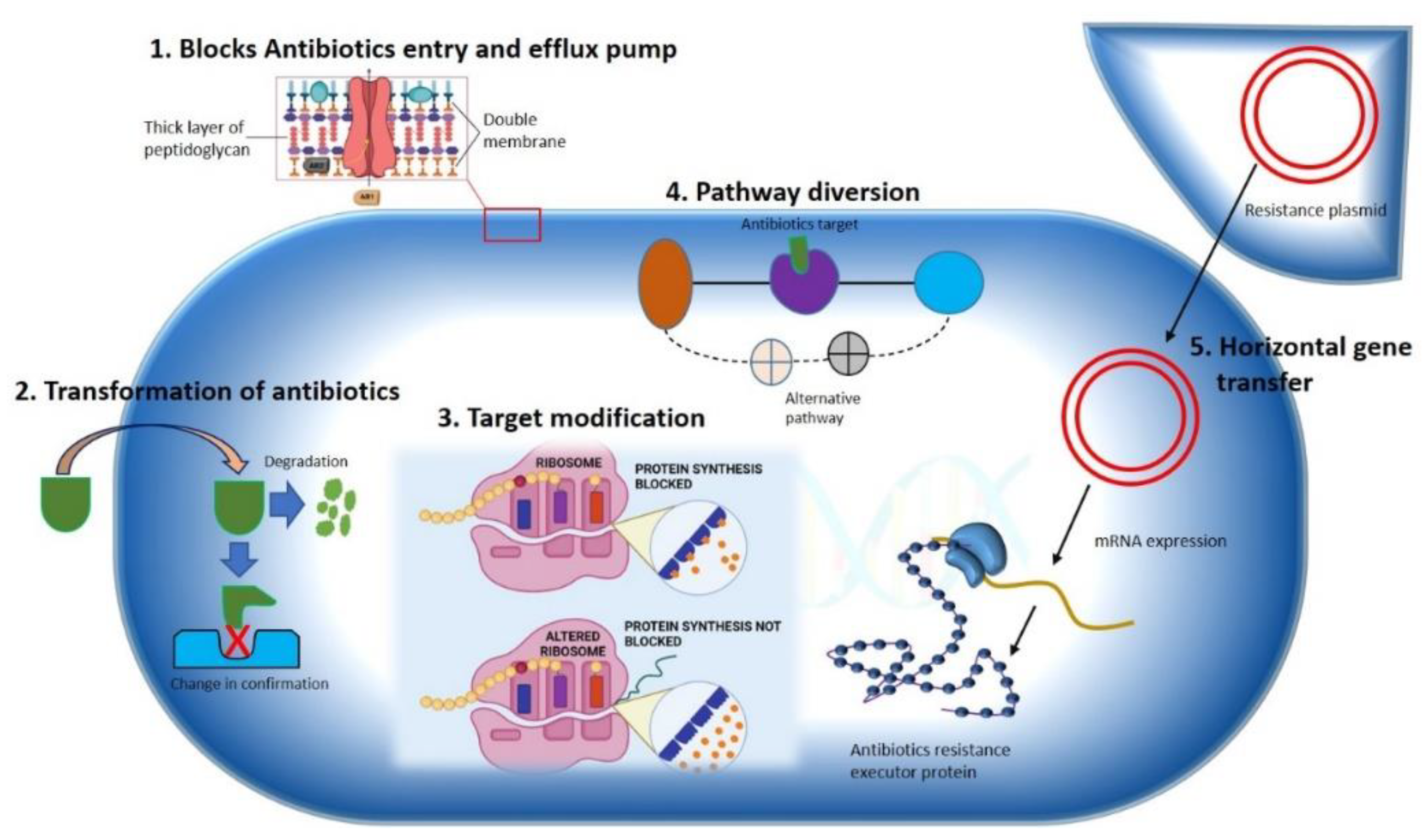

Figure 1. Mechanisms illustrating antibiotics resistance strategy used by bacteria. A Gram-negative bacterium is depicted various mechanisms for antibiotics resistance approaches against different antimicrobial agents are specified. (1) Hindering of antibiotics’ entry and removal using efflux pump. (2) Transformation of drug either by degradation or by changing drugs’ original confirmation. (3) Modification of antibiotics’ target by gene mutation. (4) Diversion of pathway containing drug target. (5) Horizontal transfer of gene.
2.1. Constrained Antibiotic Entry and Efflux Pumps
Phylogenic analysis of membrane protein signature sequence demonstrates that Gram-negative (diderms) bacteria possess bilayer membrane (outer membrane and inner membrane coverings) while another division of organisms exists with simple monolayer covering (monoderms) [7]. The outer membrane of Gram-negative bacteria contains some classical porins that are tightly regulated by certain genes and can be influenced by environmental factors [7], allowing minute molecules like amino acids and saccharides to pass through [8]. To access the specific target, this complex outer membrane of Gram-negative bacteria must be passed by most the antibacterial agents, for example, utilizing diffusion mechanism hydrophobic drugs can make an entry into the bacterial cell, hydrophilic antibiotics such as β-lactam pass through porins, while vancomycin gets hindered due to its structure. On the other hand, Gram-positive bacteria lack this complex membrane structure, making Gram-negative bacteria more antibiotic resistant than Gram-positive due to their outer membrane′s selective permeability [9]. Bacterial species that lack targets for specific antibiotics are naturally resistant to that antibiotic class. Some bacteria are naturally lacking in the targets of some antibiotics groups, which makes them intrinsically resistant to these antibiotics. Mycoplasma spp., for example, has a cell wall resistant to the cell wall synthesis inhibitor antibiotics, such as β-lactam and glycopeptides [10].
A bacterial efflux pump is another crucial mechanism for antibiotic resistance. The efflux pumps are found naturally in some bacteria or can be obtained from external sources. In bacteria, five genes that encode different efflux pumps are the major facilitator superfamily (MFS), ABC (ATP binding cassette) MDR transporter, resistance nodulation division (RND) family, staphylococcal multiresistance (SMR), and multidrug and toxic compound extrusion (MATE) families [11]. RND family proteins are found in the Gram-negative enteric bacteria E. coli and include inner membrane transporter efflux (e.g., AcrB), outer membrane protein channel (e.g., TolC), and a periplasmic accessory protein (e.g., AcrA). Numerous bacterial species such as E. coli, P. aeruginosa, Campylobacter jejuni, and Neisseria gonorrhoeae share a high homologous efflux pump such as AcrB/AcrB, mexB/MexB, B/CmeB, and mtrD/Mtrd [12]. Gram-positive bacteria, on the other hand, have only two types of efflux pumps (PmrA and NorA), both are present in S. aureus and S. pneumoniae and belong to the MFS family. Another MATE MDR efflux pump has been found in Gram-positive and Gram-negative bacteria. It uses PMF and the sodium ion gradient as an energy source, enabling the organism to develop resistance to a variety of antibiotics. Tetracycline resistance genes (63 tet genes reported) are the most common acquired resistance genes in both Gram-positive and Gram-negative clinical isolates, which encodes numerous efflux transporters [13]. Tetracycline resistance gene tet(63), which serves as an efflux pump, has been found in a multiresistance plasmid from S. aureus chicken isolates [14].
2.2. Transformation or Termination of Antibiotics
The modification and termination of antibiotics either by chemical alteration or molecular destruction is the most common approach used by bacteria to render antibiotics, making them futile with the use of enzymes. The enzymes involved in drug modification are categorized into three primary groups based on their reaction mechanisms: hydrolases, transferases, and oxidoreductases. β-lactamase is the important member of the group hydrolases, which catalyzes the hydrolytic breakdown of β-lactam ring present in cell wall synthesis inhibitor antibiotics groups such as penicillins, cephalosporins, carbapenems, monobactams, and clavulanate. A total of four different types of β-lactamase enzymes have been reported. Among them, extended-spectrum β-lactamase is the most potent enzyme which can inactivate the β-lactam antibiotics, including all penicillins, third-generation cephalosporins, and monobactam aztreonam [15] which became a challenge for the treatment of nosocomial infections [16]. Aminoglycoside-modifying enzymes (AMEs) are transferase enzymes that are found in most mobile genetic elements (MGEs), that facilitate the rapid spreading of genes. The clinically relevant member of the AMEs family is N-acetyltransferase, which acetylates aminoglycosides rather than O-phosphotransferases and O-adenylyltransferases, which adenylate and phosphorylate aminoglycosides in several locations, rendering antibiotics ineffective to its targets [17]. These antibiotic resistance genes are naturally expressed in antibiotic producing organisms such as species of Streptomyces like S. griseus, which converts streptomycin into inactive precursor streptomycin -6-phosphate. The presence of such genes in antibiotic producing organisms is still up for dispute as to whether they play a role in antibiotic resistance or only serve as housekeeping genes [18]. However, studies of these genes and their pattern could be helpful for the investigation of genetic information presents in MGEs found in various hospital isolates contributing modification of different lifesaving antibiotics.
2.3. Pathway Alteration to Avoid the Antibiotic Target Site
Bacteria commonly exploit alteration or skipping of the drug target to acquire resistance to specific antibiotics. Bacteria can evolve new metabolic sites that perform the same biochemical functions as the original target but are not inhibited by medications that target the original target. Antibiotics in the sulfa group target the bacterial folic acid synthesis pathway, which is missing in humans and higher eukaryotes. Eukaryotes get their folic acid from external sources, and they have a folate uptake system that most prokaryotes lack. Sulphonamide antibiotics primarily block dihydropteroate synthase (DHPS), a folic acid biosynthesis pathway enzyme absent in eukaryotes. Clinical isolates of some gut bacteria that have acquired a resistant variant of the DPHS gene by horizontal gene transfer have been reported to be resistant to sulphonamides (Figure 2). A unique DHPS enzyme variation generated from the genes sul1 and sul2 have been discovered in most sulphonamide-resistant enteric bacteria, which is sulphonamide insensitive but has equal binding efficacy with its substrate p-aminobenzoic acid [19,20][19][20]. This sulphonamide resistance mechanism is an excellent example of a pathway bypass resistant mechanism that avoids the antibiotic target. Other examples of pathway alteration mechanisms are the acquisition of external penicillin-binding protein (PBP2a) in MRSA clinical isolates and cluster van-genes in vancomycin-resistant enterococci. In both instances, the acquired modified enzymes functioned normally for cell wall synthesis even in the presence of cell wall synthesis inhibitor antibiotics. The mecA gene in S. aureus, which was probably obtained from Sthaphylococcus scSthaphylococcus sciuriuri, encodes an alternative PBP2a enzyme that has a poor affinity for most β-lactam antibiotics, such as penicillins, cephalosporins, and carbapenems [21]. The acquired gene mecA placed into a large MGE gene cassette, [22], providing an alternative way to bypass the target of all β-lactam antibiotics, making the organism clinically more virulent than the wild type. The van genes (vanA, vanB, vanC, vanD, vanE, vanF, vanG, vanL, vanM, and van N) encode a biochemical system in enterococci that synthesizes D-Ala rather than D-Lac or D-serine (low resistance) and destroys the wild type “D-Ala-D-Ala” ending of peptidoglycan precursor molecules, lowering vancomycin affinity by thousands of times. A clinically contagious vancomycin resistant enterococci strain can be developed by acquiring a single cluster of van gene.
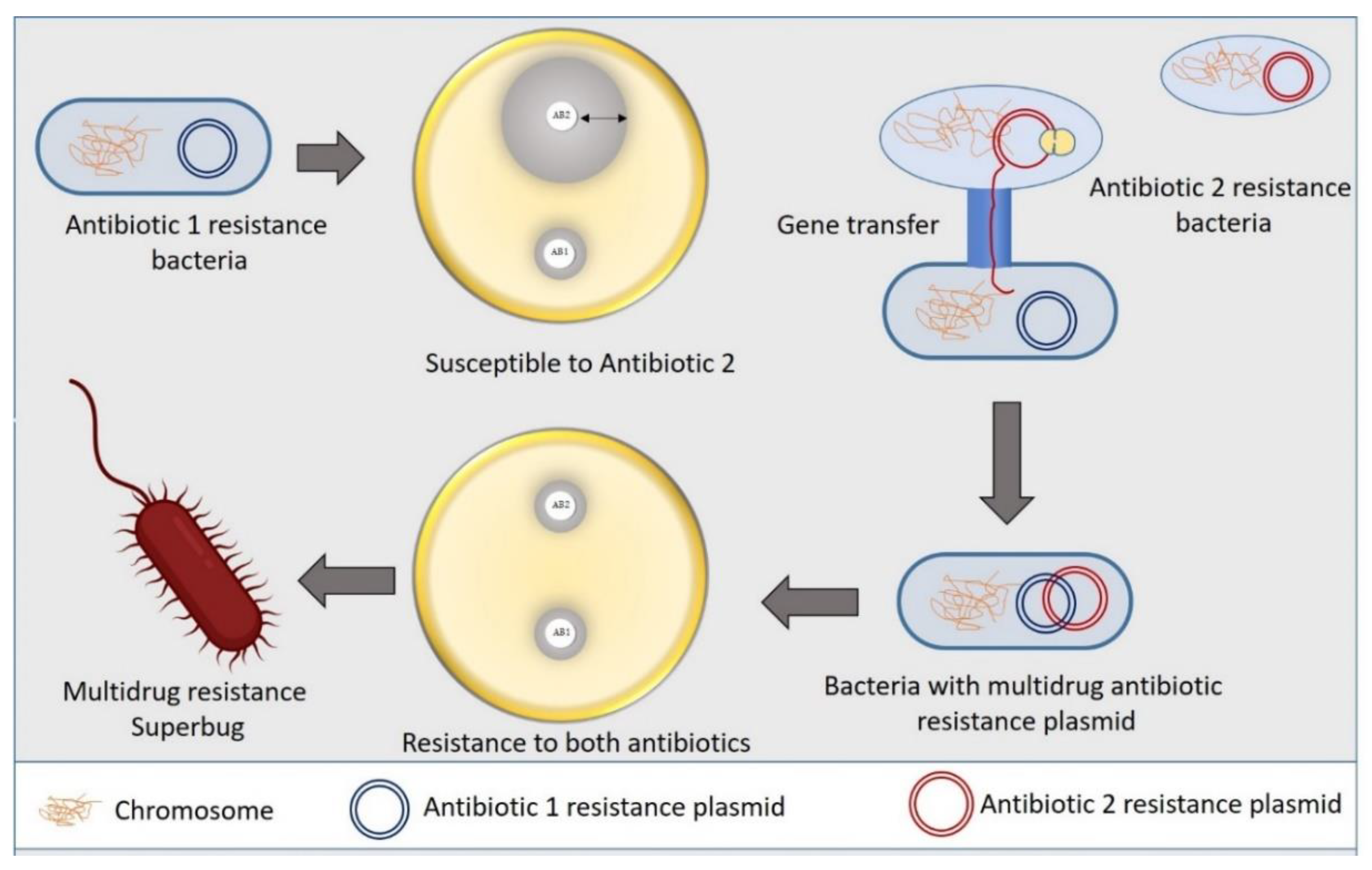

Figure 2. Mechanism of bacterial superbug formation by acquiring multiple drug resistance plasmid through horizontal gene transfer.
Bacteria can also adapt an alternative metabolic mechanism termed as a metabolic adaptation to resist some drugs. By metabolic adaptation, the daptomycin challenge in S. aureus redirects carbon flow from the TCA cycle into the pentose phosphate pathway, increasing the essential intermediates required for peptidoglycan biosynthesis intermediates, teichoic acids, and nucleosides, resulting in the development of a daptomycin non-susceptible strain from a susceptible strain [23]. Furthermore, when a daptomycin sensitive strain of S. mitis was exposed to daptomycin, the glucose metabolic pathway was transformed, resulting in the formation of a new daptomycin nonsusceptible strain [24]. The development of an alternative metabolic pathway to survive in a hostile environment containing antibiotics and the immune system of the host, makes bacteria more virulent.
2.4. Antibiotics Target Modification
Bacteria gain resistance to a wide spectrum of antibiotics by modifying target enzymes, either through genetic mutations that encode target enzymes or by modifying enzyme confirmations. Most bacteria acquire point mutations for genetic alteration, while phosphorylation, acetylation, and adenylation are the most common ways of modulating enzymatic confirmation. In both scenarios, a modified enzyme is formed that seems to have a low affinity for antibiotics while maintaining a wild-type affinity for its substrate. Rifamycin (RIF) antibiotics target DNA-dependent RNA polymerase by binding in the β-subunit RIF binding pocket, obstructing the pathway of nascent RNA. One step point mutation in the ropB (RNA polymerase) gene results in mutational resistance to RIF antibiotics. A single-step mutation substitutes one amino acid, decreasing drug affinity while maintaining the enzyme′s polymerase catalytic activity, allowing for RNA production similar to that of the wild type [25,26][25][26].
Another mechanism of bacteria to confer target modification is by enzymatic reform of drug target. The illustration of enzymatic alteration of the antibiotic target is best implied in macrolides resistance accomplished by the methylation genes to the target of macrolides, 23S rRNA ribosomal subunit [27]. The erythromycin ribosomal methylation (erm) gene produces adenine methylation at the A2058 position of the 50S ribosomal subunit′s 23S rRNA, V domain. More than 30 types of erm genes have been identified in MGEs, which may be responsible for the distribution of erm genes among different types of aerobic, anaerobic, and both Gram-positive and Gram-negative bacterial populations. Similarly, cfr genes encoding a wide range of methylase family members have been found in S. aureus, E. feacalis, E. faceium, and a few Gram-negative isolates that have been integrated into MGEs plasmids, conferring resistance to phenicols, lincosamides, and streptogramin-A. [28].
3. Mechanism of Advanced Nanotechnological Approaches against Antibiotic Resistance
Recently, various drug-loaded nanoparticles have made remarkable progress with their promising antibacterial activity in a wide range of bacteria. They offer greater protection and combat the multidrug resistance against these micro-organisms. For example, silver nanoparticles (Ag-NPs) exert robust, broad-spectrum antibacterial efficacy by generating reactive oxygen species (ROS) to kill bacteria [29]. In thHe following section, we have discussedre is various mechanisms/pathways targeted by nanoparticles to control bacterial growth or exert bactericidal activity (Figure 3).
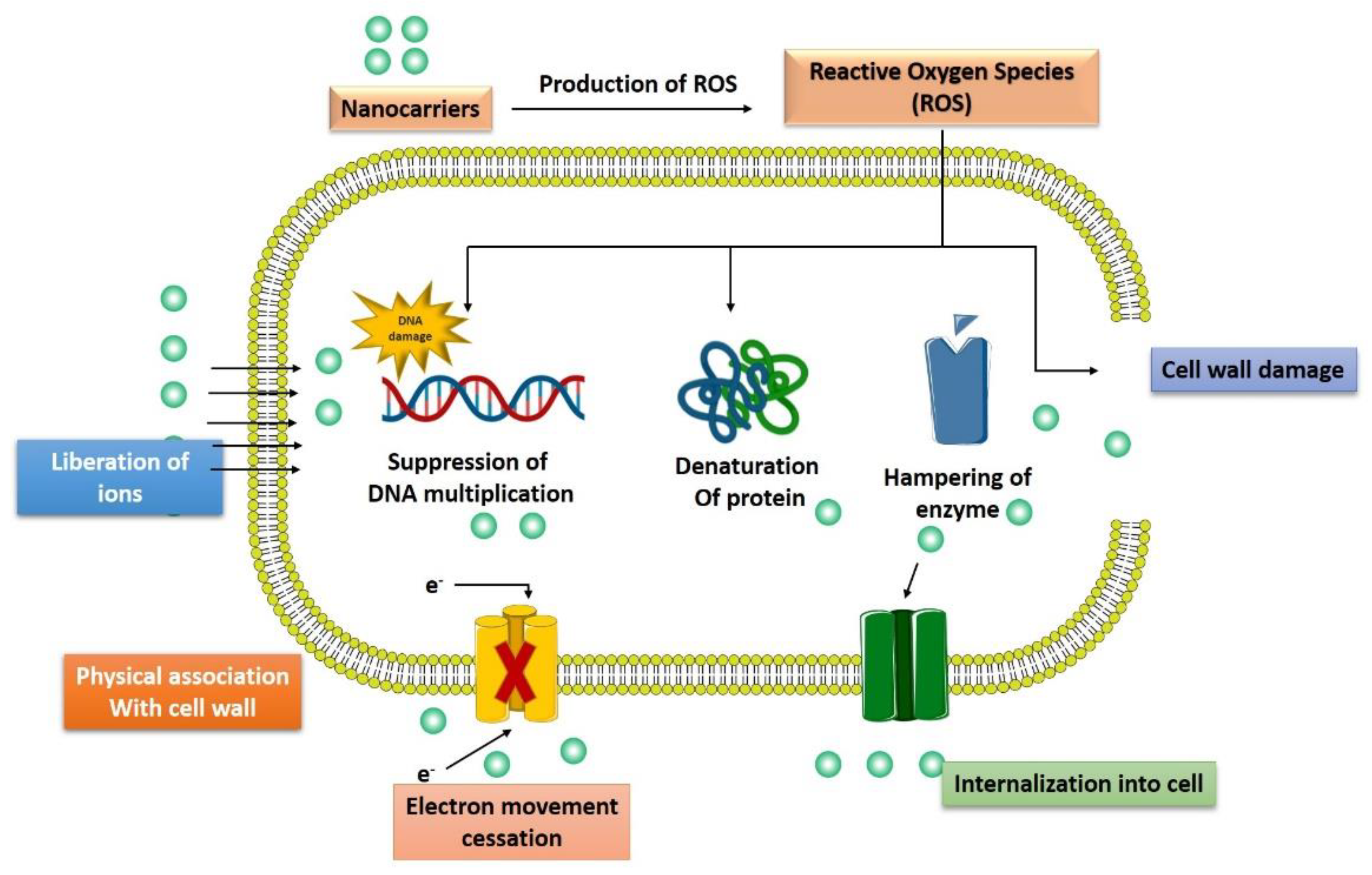

Figure 3.
Multiple pathways followed by advanced nanotechnological approaches to combat the antibiotic resistance.
3.1. Interaction with the Bacterial Cell Wall
The bacterial cell wall is a crucial defensive barrier that allows bacterial to resist external insults and maintain their natural morphology. Among various components of the cell wall, lipopolysaccharide (LPS) is particularly found on Gram-negative bacterial, while teichoic acid is found in Gram-positive bacteria [30]. Comparatively, nanoparticles are found to be more efficacious against Gram-positive than Gram-negative bacteria because LPS and phospholipid in Gram-negative bacteria form a barrier against penetration of nanoparticles and macromolecules while teichoic acid and peptidoglycan along with numerous pores in Gram-positive allows entrance of foreign particles including nanoparticles/macromolecules [31]. A novel nanoparticles of hydroxyapatite whisker/nano zinc oxide was synthesized by Yu and associates demonstrated the better antimicrobial effect of these nanoparticles on different types of bacteria such as S. mutans, Candida albicans, and S. aureus compared to E. coli. The bactericidal activity of nanoparticles was dependent on (1) the composition of a bacterial cell that might results in better activity against Gram-positive bacteria; (2) the specific parts of Gram-negative bacteria (for example LPS) that stop the binding of nanoparticles to the bacterial cell wall and modulate the endo and exocytosis of ions from bacterial cell membrane. (3) the dimensions of the bacterial cell wall (20–80 nm in Gram-positive and 1.5–10 nm in Gram-negative bacteria) affect the biological activity of nanoparticles [32]. Another study observed that phospholipid groups in the LPS layer of E. coli (Gram-negative) intermingle with ε-poly-l-lysine mediated by electrostatic attraction, resulting in the damage of the cell membrane. Nevertheless, in the case of Listeria innocua (Gram-positive), the film consists of lysine-derived phospholipids which is amphoteric in nature. Therefore, it lacks enough negative charge to attract cationic peptides. Thus, the cell membrane of L. innocua has lower permeability than that of E. coli [33]. Another study investigating the antibacterial potency of nanodiamonds found that these nanodiamonds form covalent bonds with proteins and other molecules on bacterial cell walls. The binding of nanodiamonds with bacterial intracellular elements could further impair the crucial enzymes and proteins, ultimately hindering the bacterial metabolism leading to cell death [34].
3.2. Inhibition of Biofilm Formations
Bacterial biofilm is a matrix of extracellular polymeric components around bacteria colonies that renders them resistant to antibiotic therapy, chemical disinfection, immune response [35]. Nanoparticles such as zinc oxide nanoparticles (ZnO NPs) are already established as a potent antibiofilm formulation. Kaur et al., 2020 showed that ZnO NPs inhibit biofilm in the range of bacteria and alter the bacterial cell membrane permeability [36]. Similarly, synthetic drugs or natural compound-loaded nanoparticles are potent inhibitors of bacterial biofilm. Curcumin-loaded chitosan nanoparticles exhibited robust antibiofilm activity against Candida albicans and Staphylococcus aureus. This nanoparticle not only reduced the thickness of biofilm but also kills the bacteria [37]. Similarly, synthetic antibiotics such as ciprofloxacin-loaded polylactic acid-glycolic acid (PLGA) nanospheres [38], azithromycin-loaded PAMAM-AZM nanoparticles [39], cefotaxime-loaded chitosan nanocarriers [40] can reduce >95% of Pseudomonas aeruginosa biofilm while roxithromycin-loaded cyclodextrin nanocarriers [41] and trifluorosan-loaded micellar nanocarriers [42] and vancomycin-loaded PGLA nanospheres [43] significantly inhibited the Staphylococcus aureus biofilm resulting in bacterial death.
3.3. Activation of Innate as Well as Adaptive Host Immune Response
Various investigations have revealed the interaction of nanoparticle formulation with the innate immune system leading to immune activation, and these are mediated by immune cells such as monocytes, macrophages, neutrophils, and other cells that express pattern recognition receptors (PRR). These cells release several cytokines and chemokines as a defensive mechanism against invading pathogens, including bacteria [44]. Some nanoformulation can amplify the PRR-driven inflammation, for example, the production of interleukin-1β (IL-1β), while another nanoformulation amplifies the effects of the bacterial stimulation. IL-1 signaling triggered by Streptococcus suis serotype 2 leads to the clearance of bacteria and inflammation of systemic disease as revealed by the mice model of infection [45]. Some bacterial strain for example, Staphylococcus aureus have developed an immune system bypass mechanism by releasing biofilms that lead to a significant reduction in the detection of any immune response [46]. The host innate immune system activation in response to bacterial infection primarily occurs via the identification of a pattern of bacteria surface by PRRs, such as toll-like receptors. Moreover, microorganisms that invade the cell are identified by a family of cytosolic PRRs called NOD-like receptors (NLRs). The pivotal function of the host innate immune response is to halt bacterial development and spread. In contrast, impairment of immune responses, for example, chronic wounds of diabetes mellitus patients, provides a favorable environment for bacterial for excessive proliferation leading to delay in wound healing and further tissue damage/infection [47]. Nanoparticles can impact the innate immune system response against bacteria in various ways, such as (1) nanoparticles coating the bacterial surface can result in inhibition of cellular uptake of bacteria; (2) nanoparticles (less than 30 nm size) coating bacteria can also conceal surface pathogen-associated molecular patterns (PAMPs) and activate PRR; (3) nanoparticles may compete with the process of phagocytosis of bacteria; (4) nanoparticles may induce alteration in the epigenetic pathway and (5) cytotoxic nanoparticles result in altered cell membrane permeability that becomes favorable for bacterial influx. Nanoparticles coating bacterial surfaces can serve multiple purposes, such as inhibition of bacterial pathogenicity, invasion, and concealing the PAMPs to which TLRs or NLRs would otherwise bind [48]. The pretreatment of mice macrophage with gold or silica nanoparticles can inhibit phagocytosis of killed E. coli [49], while superparamagnetic iron oxide nanoparticles can inhibit the uptake of killed S. pneumoniae by macrophage derived from bone marrow [50]. Likewise, pretreatment with gold nanoparticles to human monocytes reduces the immune activity of monocytes to live Bacilli Calmette-Guérin as seen by the level of cytokine production [51].
3.4. Generation of Reactive Oxygen Species
The toxic effect caused by various nanoparticles, such as silver nanoparticles, is due to the generation of ROS, impairment in antioxidant enzyme level, and production of free radicals, such as superoxide anions, hydrogen peroxide, hydroxyl radical, singlet oxygen, and hypochlorous acid [52]. Oxidative stress is harmful to both bacteria as well as the human body as it results in the progression of various diseases [53]. Ag-NPs can provide an alternative approach for multidrug-resistant bacterial strains such as S. aureus and P. aeruginosa. A study on these two bacterial strains isolated from breast inflamed goats found that Ag-NPs exert promising antibacterial activity via production of ROS, malondialdehyde (a marker of oxidative stress), and loss of essential components such as proteins and sugars due to leakage in the cells of the bacteria. The Ag-NPs also downregulated the initiation of antioxidant enzyme; glutathione, superoxide dismutase, and catalase [54]. Similarly, zinc oxide nanoparticle (ZnO NPs) is another promising formulation that results in an oxidative stress-mediated genotoxic effect in a radiation-resistant bacterial strain, Deinococcus radiodurans. A study by Singh et al., 2020 demonstrated that ZnO NPs were internalized remarkably inside the D. radiodurans and these nanoparticles initiate significant ROS generation, oxidation of protein molecules, and DNA impairment along with depletion in the thiol levels [55]. Similarly, copper-maleamate-functionalized mesoporous silica nanoparticles and cadmium oxide nanoparticles [56] were also found to possess potent antibacterial activity mediated by induction of oxidative stress.
3.5. Induction of Intracellular Effects
Nanoparticles can interfere with a bacterial intracellular function such as protein synthesis, enzyme function, or impair the genetic material and kill them. ZnO NPs were found to reduce the expression of various DNA overhaul genes (Mut S, Mut L, Ung, Mut M, DNA polymerase, and DNA ligase) and metabolic pathway genes (aconitase, succinate dehydrogenase) and increase the expression of DNA impairment response genes (Ddr A, Ddr B, Ddr D) [55]. Carbonate-coated Ag-NPs bind to the E. coli proteins such as tryptophanase and results in loss of enzyme activity [57]. Silver (Ag) ions are recognized to bind to the DNA of bacteria and it was revealed that these silver ions exposed to E. coli and S. aureus result in condensed DNA, ultimately arresting bacterial multiplication [58]. The interaction of Ag-NPs to E. coli also results in the upregulation of 161 genes and downregulation of 27 genes [59]. The upregulated genes were associated with a number of functions such as the citric acid cycle (sdhC), protein efflux (fsr, yajR, emrE), membrane structure and biofilm formation (bolA), electron transfer (sdhC), cellular transport (mdfA), and DNA repair (recN, uvrA, ybfE, yebG, ssb, sbmc, and nfo) [60].
References
- Gandra, S.; Tseng, K.K.; Arora, A.; Bhowmik, B.; Robinson, M.L.; Panigrahi, B.; Laxminarayan, R.; Klein, E.Y. The Mortality Burden of Multidrug-resistant Pathogens in India: A Retrospective, Observational Study. Clin. Infect. Dis. 2019, 69, 563–570.
- Gao, W.; Thamphiwatana, S.; Angsantikul, P.; Zhang, L. Nanoparticle approaches against bacterial infections. WIREs Nanomed. Nanobiotechnology 2014, 6, 532–547.
- Jiang, L.; Lin, J.; Taggart, C.; Bengoechea, J.; Scott, C.J. Nanodelivery strategies for the treatment of multidrug-resistant bacterial infections. J. Interdiscip. Nanomed. 2018, 3, 111–121.
- Chia, P.Y.; Sengupta, S.; Kukreja, A.; Ponnampalavanar, S.S.; Ng, O.T.; Marimuthu, K. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrob. Resist. Infect. Control 2020, 9, 1–11.
- Nnadozie, C.F.; Odume, O.N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 2019, 254, 113067.
- Salyers, A.; Shoemaker, N.B. Reservoirs of Antibiotic Resistance Genes. Anim. Biotechnol. 2006, 17, 137–146.
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656.
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340.
- Miller, S.I. Antibiotic Resistance and Regulation of the Gram-Negative Bacterial Outer Membrane Barrier by Host Innate Immune Molecules. mBio 2016, 7, e01541–e01616.
- van Duijkeren, E.; Schink, A.-K.; Roberts, M.C.; Wang, Y.; Schwarz, S. Mechanisms of Bacterial Resistance to Antimicrobial Agents. Microbiol. Spectr. 2018, 6, 186–198.
- Piddock, L.J.V. Clinically Relevant Chromosomally Encoded Multidrug Resistance Efflux Pumps in Bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402.
- Ma, D.; Cook, D.N.; Alberti, M.; Pon, N.G.; Nikaido, H.; Hearst, J.E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 1993, 175, 6299–6313.
- Feng, Z.; Liu, D.; Wang, L.; Wang, Y.; Zang, Z.; Liu, Z.; Song, B.; Gu, L.; Fan, Z.; Yang, S.; et al. A Putative Efflux Transporter of the ABC Family, YbhFSR, in Escherichia Coli Functions in Tetracycline Efflux and Na+(Li+)/H+ Transport. Front. Microbiol. 2020, 11, 556.
- Zhu, Y.; Wang, C.; Schwarz, S.; Liu, W.; Yang, Q.; Luan, T.; Wang, L.; Liu, S.; Zhang, W. Identification of a novel tetracycline resistance gene, tet(63), located on a multiresistance plasmid from Staphylococcus aureus. J. Antimicrob. Chemother. 2021, 76, 576–581.
- Egorov, A.M.; Ulyashova, M.M.; Rubtsova, M.Y. Bacterial Enzymes and Antibiotic Resistance. Available online: https://pubmed.ncbi.nlm.nih.gov/30713760/ (accessed on 9 November 2021).
- Collins, V.L.; Marchaim, D.; Pogue, J.M.; Moshos, J.; Bheemreddy, S.; Sunkara, B.; Shallal, A.; Chugh, N.; Eiseler, S.; Bhargava, P.; et al. Efficacy of Ertapenem for Treatment of Bloodstream Infections Caused by Extended-Spectrum-β-Lactamase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2012, 56, 2173–2177.
- Zárate, S.G.; De La Cruz Claure, M.L.; Benito-Arenas, R.; Revuelta, J.; Santana, A.G.; Bastida, A. Overcoming Aminoglycoside Enzymatic Resistance: Design of Novel Antibiotics and Inhibitors. Molecules 2018, 23, 284.
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928.
- Sköld, O. Sulfonamide resistance: Mechanisms and trends. Drug Resist. Updat. 2000, 3, 155–160.
- Rådström, P.; Swedberg, G.; Sköld, O. Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob. Agents Chemother. 1991, 35, 1840–1848.
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 464–473.
- Hiramatsu, K.; Ito, T.; Tsubakishita, S.; Sasaki, T.; Takeuchi, F.; Morimoto, Y.; Katayama, Y.; Matsuo, M.; Kuwahara-Arai, K.; Hishinuma, T.; et al. Genomic Basis for Methicillin Resistance inStaphylococcus aureus. Infect. Chemother. 2013, 45, 117–136.
- Gaupp, R.; Lei, S.; Reed, J.M.; Peisker, H.; Boyle-Vavra, S.; Bayer, A.S.; Bischoff, M.; Herrmann, M.; Daum, R.S.; Powers, R.; et al. Staphylococcus aureus Metabolic Adaptations during the Transition from a Daptomycin Susceptibility Phenotype to a Daptomycin Nonsusceptibility Phenotype. Antimicrob. Agents Chemother. 2015, 59, 4226–4238.
- Parrett, A.; Reed, J.M.; Gardner, S.G.; Mishra, N.N.; Bayer, A.S.; Powers, R.; Somerville, G.A. Metabolic changes associated with adaptive resistance to daptomycin in Streptococcus mitis-oralis. BMC Microbiol. 2020, 20, 162.
- Floss, H.G.; Yu, T.-W. RifamycinMode of Action, Resistance, and Biosynthesis. Chem. Rev. 2005, 105, 621–632.
- Campbell, E.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural Mechanism for Rifampicin Inhibition of Bacterial RNA Polymerase. Cell 2001, 104, 901–912.
- Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995, 39, 577–585.
- Marshall, S.H.; Donskey, C.J.; Hutton-Thomas, R.; Salata, R.A.; Rice, L.B. Gene Dosage and Linezolid Resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 2002, 46, 3334–3336.
- Tang, S.; Zheng, J. Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Healthc. Mater. 2018, 7, 1701503.
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249.
- Sarwar, A.; Katas, H.; Noradila Samsudin, S.; Mohamad Zin, N.; Malaysia, K.; Campus, K.L.; Abdul Aziz, J.R.M.; Lumpur, K.; Rittschof, D. Regioselective Sequential Modification of Chitosan via Azide-Alkyne Click Reaction: Synthesis, Characterization, and Antimicrobial Activity of Chitosan Derivatives and Nanoparticles. PLoS ONE 2015, 10, e0123084.
- Yu, J.; Zhang, W.; Li, Y.; Wang, G.; Yang, L.; Jin, J.; Chen, Q.; Huang, M. Synthesis, characterization, antimicrobial activity and mechanism of a novel hydroxyapatite whisker/nano zinc oxide biomaterial. Biomed. Mater. 2014, 10, 015001.
- Hyldgaard, M.; Mygind, T.; Vad, B.S.; Stenvang, M.; Otzen, D.; Meyer, R.L. The Antimicrobial Mechanism of Action of Epsilon-Poly-L-Lysine. Appl. Environ. Microbiol. 2014, 80, 7758–7770.
- Wehling, J.; Dringen, R.; Zare, R.N.; Maas, M.; Rezwan, K. Bactericidal Activity of Partially Oxidized Nanodiamonds. ACS Nano 2014, 8, 6475–6483.
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59.
- Kaur, T.; Putatunda, C.; Vyas, A.; Kumar, G. Zinc oxide nanoparticles inhibit bacterial biofilm formation via altering cell membrane permeability. Prep. Biochem. Biotechnol. 2020, 51, 309–319.
- Ma, S.; Moser, D.; Han, F.; Leonhard, M.; Schneider-Stickler, B.; Tan, Y. Preparation and antibiofilm studies of curcumin loaded chitosan nanoparticles against polymicrobial biofilms of Candida albicans and Staphylococcus aureus. Carbohydr. Polym. 2020, 241, 116254.
- Baelo, A.; Levato, R.; Julián, E.; Crespo, A.; Astola, J.; Gavaldà, J.; Engel, E.; Mateos-Timoneda, M.A.; Torrents, E. Disassembling bacterial extracellular matrix with DNase-coated nanoparticles to enhance antibiotic delivery in biofilm infections. J. Control. Release 2015, 209, 150–158.
- Gao, Y.; Wang, J.; Chai, M.; Li, X.; Deng, Y.; Jin, Q.; Ji, J. Size and Charge Adaptive Clustered Nanoparticles Targeting the Biofilm Microenvironment for Chronic Lung Infection Management. ACS Nano 2020, 14, 5686–5699.
- Jamil, B.; Habib, H.; Abbasi, S.A.; Ihsan, A.; Nasir, H.; Imran, M. Development of Cefotaxime Impregnated Chitosan as Nano-antibiotics: De Novo Strategy to Combat Biofilm Forming Multi-drug Resistant Pathogens. Front. Microbiol. 2016, 7, 330.
- Masood, F.; Yasin, T.; Bukhari, H.; Mujahid, M. Characterization and application of roxithromycin loaded cyclodextrin based nanoparticles for treatment of multidrug resistant bacteria. Mater. Sci. Eng. C 2016, 61, 1–7.
- Pascoal, S.; Liu, X.; Ly, T.; Fang, Y.; Rockliffe, N.; Paterson, S.; Shirran, S.L.; Botting, C.H.; Bailey, N.W. Rapid evolution and gene expression: A rapidly evolving Mendelian trait that silences field crickets has widespread effects on mRNA and protein expression. J. Evol. Biol. 2016, 29, 1234–1246.
- Chiang, W.-L.; Lin, T.-T.; Sureshbabu, R.; Chia, W.-T.; Hsiao, H.-C.; Liu, H.-Y.; Yang, C.-M.; Sung, H.-W. A rapid drug release system with a NIR light-activated molecular switch for dual-modality photothermal/antibiotic treatments of subcutaneous abscesses. J. Control. Release 2015, 199, 53–62.
- Swartzwelter, B.; Fux, A.; Johnson, L.; Swart, E.; Hofer, S.; Hofstätter, N.; Geppert, M.; Italiani, P.; Boraschi, D.; Duschl, A.; et al. The Impact of Nanoparticles on Innate Immune Activation by Live Bacteria. Int. J. Mol. Sci. 2020, 21, 9695.
- Lavagna, A.; Auger, J.-P.; Dumesnil, A.; Roy, D.; Girardin, S.E.; Gisch, N.; Segura, M.; Gottschalk, M. Interleukin-1 signaling induced by Streptococcus suis serotype 2 is strain-dependent and contributes to bacterial clearance and inflammation during systemic disease in a mouse model of infection. Veter Res. 2019, 50, 1–18.
- Horn, C.M.; Kielian, T. Crosstalk Between Staphylococcus aureus and Innate Immunity: Focus on Immunometabolism. Front. Immunol. 2021, 11, 3767.
- Dolgachev, V.A.; Yu, B.; Reinke, J.M.; Raghavendran, K.; Hemmila, M.R. Host susceptibility to gram-negative pneumonia after lung contusion. J. Trauma Inj. Infect. Crit. Care 2012, 72, 614–623.
- Khan, F.; Manivasagan, P.; Lee, J.-W.; Pham, D.T.N.; Oh, J.; Kim, Y.-M. Fucoidan-Stabilized Gold Nanoparticle-Mediated Biofilm Inhibition, Attenuation of Virulence and Motility Properties in Pseudomonas aeruginosa PAO1. Mar. Drugs 2019, 17, 208.
- Tyner, K.; Bancos, S.; Stevens, D. Effect of silica and gold nanoparticles on macrophage proliferation, activation markers, cytokine production, and phagocytosis in vitro. Int. J. Nanomed. 2014, 10, 183–206.
- Kodali, V.; Littke, M.H.; Tilton, S.C.; Teeguarden, J.; Shi, L.; Frevert, C.W.; Wang, W.; Pounds, J.; Thrall, B.D. Dysregulation of Macrophage Activation Profiles by Engineered Nanoparticles. ACS Nano 2013, 7, 6997–7010.
- Swartzwelter, B.J.; Barbero, F.; Verde, A.; Mangini, M.; Pirozzi, M.; De Luca, A.C.; Puntes, V.F.; Leite, L.C.C.; Italiani, P.; Boraschi, D. Gold Nanoparticles Modulate BCG-Induced Innate Immune Memory in Human Monocytes by Shifting the Memory Response towards Tolerance. Cells 2020, 9, 284.
- Vimbela, G.V.; Ngo, S.M.; Fraze, C.; Yang, L.; Stout, D.A. Antibacterial properties and toxicity from metallic nanomaterials. Int. J. Nanomed. 2017, 12, 3941–3965.
- Panth, N.; Paudel, K.R.; Parajuli, K. Reactive Oxygen Species: A Key Hallmark of Cardiovascular Disease. Adv. Med. 2016, 2016, 9152732.
- Yuan, Y.-G.; Peng, Q.-L.; Gurunathan, S. Effects of Silver Nanoparticles on Multiple Drug-Resistant Strains of Staphylococcus aureus and Pseudomonas aeruginosa from Mastitis-Infected Goats: An Alternative Approach for Antimicrobial Therapy. Int. J. Mol. Sci. 2017, 18, 569.
- Singh, R.; Cheng, S.; Singh, S. Oxidative stress-mediated genotoxic effect of zinc oxide nanoparticles on Deinococcus radiodurans. 3 Biotech 2020, 10, 66.
- Díaz-García, D.; Ardiles, P.R.; Prashar, S.; Rodríguez-Diéguez, A.; Páez, P.L.; Gómez-Ruiz, S. Preparation and Study of the Antibacterial Applications and Oxidative Stress Induction of Copper Maleamate-Functionalized Mesoporous Silica Nanoparticles. Pharmaceutics 2019, 11, 30.
- Wigginton, N.; de Titta, A.; Piccapietra, F.; Dobias, J.; Nesatyy, V.J.; Suter, M.J.-F.; Bernier-Latmani, R. Binding of Silver Nanoparticles to Bacterial Proteins Depends on Surface Modifications and Inhibits Enzymatic Activity. Environ. Sci. Technol. 2010, 44, 2163–2168.
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178.
- McQuillan, J.S.; Shaw, A. Differential gene regulation in the Ag nanoparticle and Ag+-induced silver stress response in Escherichia coli: A full transcriptomic profile. Nanotoxicology 2014, 8, 177–184.
- Gou, N.; Onnis-Hayden, A.; Gu, A.Z. Mechanistic Toxicity Assessment of Nanomaterials by Whole-Cell-Array Stress Genes Expression Analysis. Environ. Sci. Technol. 2010, 44, 5964–5970.
More
