Diagnostic tools play a pivotal role in warfare against schistosomiasis but must adapt to the endemic status and objectives of activities. With the decrease of prevalence and infection intensity of schistosomiasis in human beings and livestock, optimal methodologies with high sensitivity and absolute specificity are needed for the detection of asymptomatic cases or light infections, as well as disease surveillance to verify elimination. In comparison with the parasitological methods with relatively low sensitivity and serological techniques lacking specificity, which both had been widely used in previous control stages, the molecular detection methods based on the amplification of promising genes of the schistosome genome may pick up the baton to assist the eventual aim of elimination.
- schistosomiasis japonica
- elimination
- diagnostic tools
- molecular techniques
1. Traditional Diagnostic Tools Applied in Schistosomiasis Control in China
It cannot be exaggerated that diagnosis is the essential basis of schistosomiasis control for case identification and treatment, assessment of morbidity, and evaluation of control strategies, which are all dependent on the performance of diagnostic tests [27,39][1][2]. Two kinds of diagnostic methodologies, namely parasitological techniques mainly including KK and MHT [40[3][4],41], and immunologic approaches based on detection of specific antibodies, were widely used in the national control program in China, accelerating the process of schistosomiasis control significantly [11,42][5][6].
1.1. Parasitological Methods
1.2. Immunologic Tests
2. Molecular Methods Developed to Detect the Pathogen of Schistosomiasis
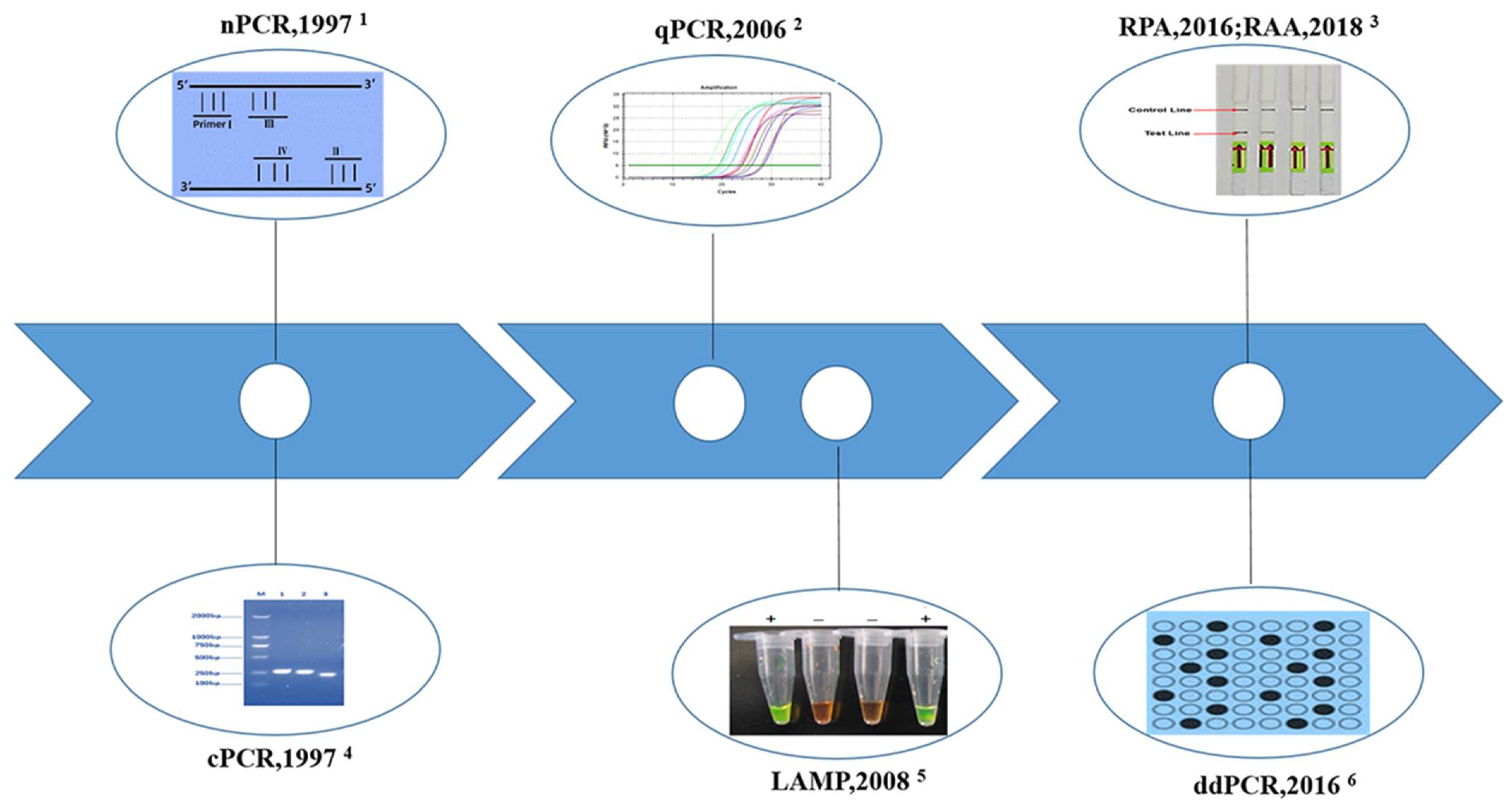
2.1. Conventional Polymerase Chain Reaction (cPCR)
2.2. Nested PCR (nPCR)
2.3. Real-Time Quantitative PCR (qPCR)
2.4. Droplet Digital PCR (ddPCR)
2.5. Loop-Mediated Isothermal Amplification (LAMP)
2.6. Recombinase Polymerase Amplification (RPA)
References
- Xu, J.; Steinman, P.; Maybe, D.; Zhou, X.N.; Lv, S.; Li, S.Z.; Peeling, R. Evolution of the national schistosomiasis control programmes in the people’s republic of china. Adv Parasitol. 2016, 92, 1–38.
- Collins, C.; Xu, J.; Tang, S. Schistosomiasis control and the health system in P.R. China. Infect. Dis. Poverty 2012, 1, 8.
- Leuenberger, A.; Nassoro, T.; Said, K.; Fenner, L.; Sikalengo, G.; Letang, E.; Montresor, A.; Zhou, X.N.; Steinmann, P.; Marti, H.; et al. Assessing stool quantities generated by three specific kato-katz thick smear templates employed in different settings. Infect. Dis. Poverty 2016, 5, 58.
- Lin, D.D.; Liu, J.X.; Liu, Y.M.; Hu, F.; Zhang, Y.Y.; Xu, J.M.; Li, J.Y.; Ji, M.J.; Bergquist, R.; Wu, G.L.; et al. Routine kato-katz technique underestimates the prevalence of Schistosoma japonicum: A case study in an endemic area of the people’s republic of China. Parasitol. Int. 2008, 57, 281–286.
- Hinz, R.; Schwarz, N.G.; Hahn, A.; Frickmann, H. Serological approaches for the diagnosis of schistosomiasis—A review. Mol. Cell Probes 2017, 31, 2–21.
- Xu, J.; Peeling, R.W.; Chen, J.X.; Wu, X.H.; Wu, Z.D.; Wang, S.P.; Feng, T.; Chen, S.H.; Li, H.; Guo, J.G.; et al. Evaluation of immunoassays for the diagnosis of Schistosoma japonicum infection using archived sera. PLoS Negl. Trop. Dis. 2011, 5, e949.
- Zhang, J.F.; Xu, J.; Bergquist, R.; Yu, L.L.; Yan, X.L.; Zhu, H.Q.; Wen, L.Y. Development and application of diagnostics in the national schistosomiasis control programme in the people’s republic of China. Adv. Parasitol. 2016, 92, 409–434.
- Bergquist, R.; Yang, G.J.; Knopp, S.; Utzinger, J.; Tanner, M. Surveillance and response: Tools and approaches for the elimination stage of neglected tropical diseases. Acta Trop. 2015, 141, 229–234.
- Xu, J.; Li, S.Z.; Zhang, L.J.; Bergquist, R.; Dang, H.; Wang, Q.; Lv, S.; Wang, T.P.; Lin, D.D.; Liu, J.B.; et al. Surveillance-based evidence: Elimination of schistosomiasis as a public health problem in the peoples’ republic of China. Infect. Dis. Poverty 2020, 9, 63.
- Xu, B.; Feng, Z.; Xu, X.J.; Hu, W. Evaluation of kato-katz technique combined with stool hatching test in diagnosis of schistosomiasis japonica. Chin. J. Schistosomiasis Control 2011, 23, 321–323. (In Chinese)
- Lamberton, P.H.; Kabatereine, N.B.; Oguttu, D.W.; Fenwick, A.; Webster, J.P. Sensitivity and specificity of multiple kato-katz thick smears and a circulating cathodic antigen test for Schistosoma mansoni diagnosis pre- and post-repeated-praziquantel treatment. PLoS Negl. Trop. Dis. 2014, 8, e3139.
- Qian, M.B.; Zhuang, S.F.; Zhu, S.Q.; Deng, X.M.; Li, Z.X.; Zhou, X.N. Improving diagnostic performance of the kato-katz method for clonorchis sinensis infection through multiple samples. Parasites Vectors 2019, 12, 336.
- Zhu, H.Q.; Xu, J.; Zhu, R.; Cao, C.-L.; Bao, Z.-P.; Yu, Q.; Zhang, L.J.; Xu, X.L.; Feng, Z.; Guo, J.G. Comparison of the miracidium hatching test and modified kato-katz method for detecting Schistosoma japonicum in low prevalence areas of China. Southeast Asian J. Trop. Med. Public Health 2014, 45, 20–25.
- Lei, Z.L.; Zhou, X.N. Eradication of schistosomiasis: A new target and a new task for the national schistosomiasis control porgramme in the People’s Republic of China. Chin. J. Schistosomiasis Control 2015, 27, 1–4. (In Chinese)
- Chen, J.; Xu, J.; Bergquist, R.; Li, S.Z.; Zhou, X.N. “Farewell to the god of plague”: The importance of political commitment towards the elimination of schistosomiasis. Trop. Med. Infect. Dis. 2018, 3, 108.
- Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature 2009, 460, 345–351.
- Weerakoon, G.K.; Gordon, C.A.; McManus, D.P. DNA diagnostics for schistosomiasis control. Trop. Med. Infect. Dis. 2018, 3, 81.
- Utzinger, J.; Becker, S.L.; van Lieshout, L.; van Dam, G.J.; Knopp, S. New diagnostic tools in schistosomiasis. Clin. Microbiol. Infect. 2015, 21, 529–542.
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273.
- Weerakoon, K.G.; Gobert, G.N.; Cai, P.; McManus, D.P. Advances in the diagnosis of human schistosomiasis. Clin. Microbiol. Rev. 2015, 28, 939–967.
- He, P.; Song, L.G.; Xie, H.; Liang, J.Y.; Yuan, D.Y.; Wu, Z.D.; Lv, Z.Y. Nucleic acid detection in the diagnosis and prevention of schistosomiasis. Infect. Dis. Poverty 2016, 5, 25.
- Chen, Y.P.; Weng, X.H. Detection DNA of Schistosoma japonicum using PCR assay. Chin. J. Infect. Dis. 1997, 15, 203–206. (In Chinese)
- Chen, Y.P.; Weng, X.H. The research of utilizing PCR to detect 5D gene of Schistosoma japonicum. Chin. J. Parasitol. Parasit. Dis. 1998, 26, 13–61. (In Chinese)
- Zhou, J.; Tao, K.H.; Li, Y.X.; Qian, W.H.; Zhang, J.H.; Wang, Y.; Zhang, Z.S. Detection of Schistosomia japonicum 5D gene by polymerase chain reaction and genechip technique. Zhonghua Liu Xing Bing Xue Za Zhi 2004, 25, 154–157. (In Chinese)
- Chen, J.H.; Wen, L.Y.; Zhang, X.Z.; Zhang, J.F.; Yu, L.L.; Hong, L.D. Development of a PCR assay for detecting Schistosoma japonicum-infected Oncomelania hupensis. Chin. J. Parasitol. Parasit. Dis. 2006, 24, 204–207. (In Chinese)
- Li, H.J.; Liang, Y.S.; Dai, J.R.; Tao, Y.H.; Wang, W.; Qu, G.L.; Wei, J.Y. Homology of 18s small subunit ribosomal RNA gene among species and strains of schistosoma and sensitivity of PCR assay to detect single cercaria. Chin. J. Schistosomiasis Control 2008, 20, 418–422. (In Chinese)
- Wang, T.A.; Shen, H.Y.; Shen, Y.X.; Wang, W.B.; Wang, B.J.; Liang, Y.S.; Hong-Xiang, Z. Establishment of PCR assay for detection of Schistosoma japonicum miracidium in water. Chin. J. Zoonoses 2010, 26, 562–564. (In Chinese)
- Thanchomnang, T.; Tantrawatpan, C.; Intapan, P.M.; Sri-Aroon, P.; Limpanont, Y.; Limpanont, Y.; Lulitanond, V.; Janwan, P.; Sanpool, O.; Tourtip, S.; et al. Pyrosequencing for rapid molecular identification of Schistosoma japonicum and S. Mekongi eggs and cercariae. Exp. Parasitol. 2013, 135, 148–152.
- Kumagai, T.; Furushima-Shimogawara, R.; Ohmae, H.; Wang, T.P.; Lu, S.; Chen, R.; Wen, L.; Ohta, N. Detection of early and single infections of Schistosoma japonicum in the intermediate host snail, Oncomelania hupensis, by PCR and loop-mediated isothermal amplification (LAMP) assay. Am. J. Trop. Med. Hyg. 2010, 83, 542–548.
- Sandoval, N.; Siles-Lucas, M.; Pérez-Arellano, J.L.; Carranza, C.; Puente, S.; López-Abán, J.; Muro, A. A new PCR-based approach for the specific amplification of DNA from different schistosoma species applicable to human urine samples. Parasitology 2006, 133, 581–587.
- Feng, T.; Qin, Z.Q.; Jing, X.U.; Zhou, J.; Qian, Y.J.; Zhu, H.Q.; Lv, S.; Cao, C.L.; Li, S.Z. Efficacy evaluation of a loop mediated isothermal amplification technique in detection of DNA of Schistosoma japonicum eggs in fecal samples. Chin. J. Parasitol. Parasit. Dis. 2017, 35, 230–234. (In Chinese)
- Qin, Z.Q.; Xu, J.; Feng, T.; Lv, S.; Qian, Y.J.; Zhang, L.J.; Li, Y.L.; Lv, C.; Bergquist, R.; Li, S.Z.; et al. Field evaluation of a loop-mediated isothermal amplification (lamp) platform for the detection of Schistosoma japonicum infection in Oncomelania hupensis snails. Trop. Med. Infect. Dis. 2018, 3, 124.
- Kato-Hayashi, N.; Kirinoki, M.; Iwamura, Y.; Kanazawa, T.; Kitikoon, V.; Matsuda, H.; Chigusa, Y. Identification and differentiation of human schistosomes by polymerase chain reaction. Exp. Parasitol. 2010, 124, 325–329.
- Kato-Hayashi, N.; Leonardo, L.R.; Arevalo, N.L.; Tagum, M.N.; Apin, J.; Agsolid, L.M.; Chua, J.C.; Villacorte, E.A.; Kirinoki, M.; Kikuchi, M.; et al. Detection of active schistosome infection by cell-free circulating DNA of Schistosoma japonicum in highly endemic areas in Sorsogon Province, the Philippines. Acta Trop. 2015, 141, 178–183.
- Xia, C.M.; Rong, R.; Lu, Z.X.; Shi, C.J.; Xu, J.; Zhang, H.Q.; Gong, W.; Luo, W. Schistosoma japonicum: A PCR assay for the early detection and evaluation of treatment in a rabbit model. Exp. Parasitol. 2009, 121, 175–179.
- Driscoll, A.J.; Kyle, J.L.; Remais, J. Development of a novel PCR assay capable of detecting a single Schistosoma japonicum cercaria recovered from Oncomelania hupensis. Parasitology 2005, 131, 497–500.
- Lu, Z.X.; Xu, J.; Gong, W.; Luo, W.; Zhang, H.Q.; Rong, R.; Shi, C.J.; Xia, C.M. Detection of Schistosoma japonicum DNA by polymerase chain reaction. Chin. J. Zoonoses 2007, 23, 479–483. (In Chinese)
- Fung, M.S.; Xiao, N.; Wang, S.; Carlton, E.J. Field evaluation of a PCR test for Schistosoma japonicum egg detection in low-prevalence regions of China. Am. J. Trop. Med. Hyg. 2012, 87, 1053–1058.
- Zhao, X.; Gu, K.; Zeng, Q.; Gao, L.; Cheng, D. Diagnostic value of SjR2 gene in colonic tissue from Schistosoma japonicum infected hosts. Med. Sci. Monit. 2019, 25, 427–435.
- Xu, J.; Rong, R.; Zhang, H.Q.; Shi, C.J.; Zhu, X.Q.; Xia, C.M. Sensitive and rapid detection of Schistosoma japonicum DNA by loop-mediated isothermal amplification (LAMP). Int. J. Parasitol. 2010, 40, 327–331.
- Xu, X.; Zhang, Y.; Lin, D.; Zhang, J.; Xu, J.; Liu, Y.M.; Hu, F.; Qing, X.; Xia, C.; Pan, W. Serodiagnosis of Schistosoma japonicum infection: Genome-wide identification of a protein marker, and assessment of its diagnostic validity in a field study in China. Lancet Infect. Dis. 2014, 14, 489–497.
- Chen, C.; Guo, Q.; Fu, Z.; Liu, J.; Lin, J.; Xiao, K.; Sun, P.; Cong, X.; Liu, R.; Hong, Y. Reviews and advances in diagnostic research on Schistosoma japonicum. Acta Trop. 2021, 213, 105743.
- Laha, T.; Brindley, P.J.; Smout, M.J.; Verity, C.K.; McManus, D.P.; Loukas, A. Reverse transcriptase activity and untranslated region sharing of a new RTE-like, non-long terminal repeat retrotransposon from the human blood fluke, Schistosoma japonicum. Int. J. Parasitol. 2002, 32, 1163–1174.
- Guo, J.J.; Zheng, H.J.; Xu, J.; Zhu, X.Q.; Wang, S.Y.; Xia, C.M. Sensitive and specific target sequences selected from retrotransposons of Schistosoma japonicum for the diagnosis of schistosomiasis. PLoS Negl. Trop. Dis. 2012, 6, e1579.
- Zhang, F.; Hou, M.; Chuan-Xin, Y.U.; Yang, K.; Yang, B.Y.; Zhang, W.Y.; Luo, X.F.; Chen, L.; Min-Jun, J.I. The use of nested PCR to monitor sentinel mice in aquatic areas infested with schistosomiasis japonica. J. Pathog. Biol. 2015, 10, 325–328. (In Chinese)
- Xu, J.; Duan, Z.L.; Guan, Z.X.; Wang, Y.Y.; Lin, C.; Zhang, T.T.; Zhang, H.Q.; Qian, X.; Xia, C.M. Early detection of circulating DNA of Schistosoma japonicum in sentinel mice models. Exp. Parasitol. 2017, 176, 82–88.
- Liu, A.P.; Yang, Q.L.; Guo, J.J.; Jing, X.U.; Xia, C.M. DNA detection of low-intensity Schistosoma japonicum infection by nested-PCR assay in serum of host. Suzhou Univ. J. Med. Sci. 2010, 30, 915–917. (In Chinese)
- Zhang, X.; He, C.C.; Liu, J.M.; Li, H.; Lu, K.; Fu, Z.Q.; Zhu, C.G.; Liu, Y.P.; Tong, L.B.; Zhou, D.B.; et al. Nested-PCR assay for detection of Schistosoma japonicum infection in domestic animals. Infect. Dis. Poverty 2017, 6, 86.
- Zeng, F.S.; He, L.; He, X.M.; Yang, J.; Qin, Z.Q. Nested-PCR assay for detection of Schistosoma japonicum. J. Trop. Dis. Parasitol. 2017, 15, 136–138. (In Chinese)
- Tong, Q.B.; Lu, S.H.; Wang, T.P.; Chen, R.; Lou, D.; Zhuo, M.M. Utilization nested PCR to detect the infection of Schistosoma japonicum. Acta Parasitol. Med. Entomol. Sin. 2009, 16, 203–207. (In Chinese)
- Bell, A.; Ranford-Cartwright, L. Real-time quantitative PCR in parasitology. Trends Parasitol. 2002, 18, 338.
- Williams, P.M.; Giles, T.; Tucker, A.; Winer, J.; Heid, C. Development and application of real-time quantitative PCR. Birkhäuser Boston. 1998, 1, 313–325.
- Gordon, C.A.; Gray, D.J.; Gobert, G.N.; Mcmanus, D.P. DNA amplification approaches for the diagnosis of key parasitic helminth infections of humans. Mol. Cell Probes 2011, 25, 143–152.
- Lier, T.; Simonsen, G.S.; Haaheim, H.; Hjelmevoll, S.O.; Vennervald, B.J.; Johansen, M.V. Novel real-time PCR for detection of Schistosoma japonicum in stool. Southeast Asian J. Trop. Med. Public Health 2006, 37, 257–264.
- Lier, T.; Simonsen, G.S.; Wang, T.; Lu, D.; Haukland, H.H.; Vennervald, B.J.; Hegstad, J.; Johansen, M.V. Real-time polymerase chain reaction for detection of low-intensity Schistosoma japonicum infections in China. Am. J. Trop. Med. Hyg. 2009, 81, 428–432.
- He, P.; Gordon, C.A.; Williams, G.M.; Li, Y.; Wang, Y.; Hu, J.; Gray, D.J.; Ross, A.G.; Harn, D.; McManus, D.P. Real-time PCR diagnosis of Schistosoma japonicum in low transmission areas of china. Infect. Dis. Poverty 2018, 7, 8.
- Wu, H.W.; Qin, Y.F.; Chu, K.; Meng, R.; Liu, Y.; McGarvey, S.T.; Olveda, R.; Acosta, L.; Ji, M.J.; Fernandez, T.; et al. High prevalence of Schistosoma japonicum infection in water buffaloes in the Philippines assessed by real-time polymerase chain reaction. Am. J. Trop. Med. Hyg. 2010, 82, 646–652.
- Gordon, C.A.; Acosta, L.P.; Gray, D.J.; Olveda, R.M.; Jarilla, B.; Gobert, G.N.; Ross, A.G.; McManus, D.P. High prevalence of Schistosoma japonicum infection in carabao from samar province, the philippines: Implications for transmission and control. PLoS Negl. Trop. Dis. 2012, 6, e1778.
- Gordon, C.A.; Acosta, L.P.; Gobert, G.N.; Jiz, M.; Olveda, R.M.; Ross, A.G.; Gray, D.J.; Williams, G.M.; Harn, D.; Li, Y.; et al. High prevalence of Schistosoma japonicum and fasciola gigantica in bovines from northern samar, the philippines. PLoS Negl. Trop. Dis. 2015, 9, e0003108.
- Gordon, C.A.; Acosta, L.P.; Gobert, G.N.; Olveda, R.M.; Ross, A.G.; Williams, G.M.; Gray, D.J.; Harn, D.; Li, Y.; McManus, D.P. Real-time PCR demonstrates high prevalence of Schistosoma japonicum in the philippines: Implications for surveillance and control. PLoS Negl. Trop. Dis. 2015, 9, e0003483.
- Gordon, C.A.; McManus, D.P.; Acosta, L.P.; Olveda, R.M.; Williams, G.M.; Ross, A.G.; Gray, D.J.; Gobert, G.N. Multiplex real-time PCR monitoring of intestinal helminths in humans reveals widespread polyparasitism in northern samar, the Philippines. Int. J. Parasitol. 2015, 45, 477–483.
- Lier, T.; Johansen, M.V.; Hjelmevoll, S.O.; Vennervald, B.J.; Simonsen, G.S. Real-time PCR for detection of low intensity Schistosoma japonicum infections in a pig model. Acta Trop. 2008, 105, 74–80.
- Qin, Y.F.; Liu, Y.; Xue-Li, D.U.; Meng, R.; Chu, K.; Min-Jun, J.I.; Kurtis, J.D.; Hai-Wei, W.U. Sensitivity of SYBR green real-time copro-PCR on detection of Schistosoma japonicum infection. J. Pathog. Biol. 2009, 4, 432–435. (In Chinese)
- Van Dorssen, C.F.; Gordon, C.A.; Li, Y.; Williams, G.M.; Wang, Y.; Luo, Z.; Gobert, G.N.; You, H.; McManus, D.P.; Gray, D.J. Rodents, goats and dogs—Their potential roles in the transmission of schistosomiasis in China. Parasitology 2017, 144, 1633–1642.
- Dang-Trinh, M.A.; Angeles, J.M.M.; Moendeg, K.J.; Macalanda, A.M.C.; Higuchi, L.; Oto, C.; Kirinoki, M.; Chigusa, Y.; Kawazu, S.I. Utilization of real time PCR for the assessment of egg burden in the organs of Schistosoma japonicum experimentally infected mice. Exp. Parasitol. 2018, 189, 61–65.
- Fornillos, R.J.C.; Sato, M.A.O.; Tabios, I.K.B.; Sato, M.; Leonardo, L.R.; Chigusa, Y.; Minamoto, T.A.O.; Kikuchi, M.; Legaspi, E.R.; Fontanilla, I.K.C. Detection of Schistosoma japonicum and Oncomelania hupensis quadrasi environmental DNA and its potential utility to schistosomiasis japonica surveillance in the Philippines. PLoS ONE 2019, 14, e0224617.
- Li, Z.; Liang, B.; Zhao, Y.Y.; Huang, L.; Wang, Y.F. Fluorescent quantitative real-time PCR for detection of Schistosoma japonicum. Chin. J. Parasitol. Parasit. Dis. 2008, 26, 299–303. (In Chinese)
- Zhou, L.; Tang, J.; Zhao, Y.; Gong, R.; Lu, X.; Gong, L.; Wang, Y. A highly sensitive Taqman real-time PCR assay for early detection of schistosoma species. Acta Trop. 2011, 120, 88–94.
- Wang, B.J.; Wang, W.B.; Zhou, X.; Chen, Y.Q.; Zhang, J.; Liu, C.C.; Liang, Y.S.; Hong-Xiang, Z. Fluorescent quantitative real-time PCR for quantitative detection of Schistosoma japonicum cercariae in water. Chin. J. Zoonoses 2011, 27, 1075–1081. (In Chinese)
- Kongklieng, A.; Kaewkong, W.; Intapan, P.M.; Sanpool, O.; Janwan, P.; Thanchomnang, T.; Lulitanond, V.; Sri-Aroon, P.; Limpanont, Y.; Maleewong, W. Molecular differentiation of Schistosoma japonicum and Schistosoma mekongi by real-time PCR with high resolution melting analysis. Korean J. Parasitol. 2013, 51, 651–656.
- Li, J.; Zhao, G.H.; Lin, R.; Blair, D.; Sugiyama, H.; Zhu, X.Q. Rapid detection and identification of four major schistosoma species by high-resolution melt (HRM) analysis. Parasitol. Res. 2015, 114, 4225–4232.
- Thanchomnang, T.; Intapan, P.; Sri-Aroon, P.; Lulitanond, V.; Janwan, P.; Sanpool, O.; Maleewong, W. Molecular detection of Schistosoma japonicum in infected snails and mouse faeces using a real-time PCR assay with fret hybridisation probes. Mem. Inst. Oswaldo Cruz 2011, 106, 831–836.
- Guan, W.; Jing, X.; Liang, S.; Sun, H.; Dong, L.L.; Xia, C.M. Quantificational detection of Schistosoma japonicum DNA in serum of the host and assessment of infectiosity by real-time pcr. Chin. J. Zoonoses 2014, 30, 263–267. (In Chinese)
- Xu, M.Z.; Wen, L.; Shen, X.J.; Liao, Y.; Tian, B. RT-PCR was used to monitor Schistosoma japonicum cercariae on the water surface in the reservoir area of Xiangjiang Changsha comprehensive hub project from 2016 to 2017. Pract. Prev. Med. 2021, 28, 837–839. (In Chinese)
- Guo, Q.; Chen, C.; Zhou, K.; Li, Y.; Tong, L.; Yue, Y.; Zhou, K.; Liu, J.; Fu, Z.; Lin, J.; et al. Evaluation of a real-time pcr assay for diagnosis of schistosomiasis japonica in the domestic goat. Parasites Vectors 2020, 13, 535.
- Hung, Y.W.; Remais, J. Quantitative detection of Schistosoma japonicum cercariae in water by real-time PCR. PLoS Negl. Trop. Dis. 2008, 2, e337.
- Worrell, C.; Xiao, N.; Vidal, J.E.; Chen, L.; Zhong, B.; Remais, J. Field detection of Schistosoma japonicum cercariae in environmental water samples by quantitative PCR. Appl. Environ. Microbiol. 2011, 77, 2192–2195.
- Ben-Jing, W.; Wen-Bo, W.; Xia, Z.; You-Sheng, L.; Hong-Xiang, Z. Establishment eraly diganostic method for Schistosoma japonicum infetion based on mice model. Beijing Forum Trop. Med. Parasitol. 2011. (In Chinese)
- Pomari, E.; Piubelli, C.; Perandin, F.; Bisoffi, Z. Digital PCR: A new technology for diagnosis of parasitic infections. Clin. Microbiol. Infect. 2019, 25, 1510–1516.
- Weerakoon, K.G.; Gordon, C.A.; Gobert, G.N.; Cai, P.; McManus, D.P. Optimisation of a droplet digital PCR assay for the diagnosis of Schistosoma japonicum infection: A duplex approach with DNA binding dye chemistry. J. Microbiol. Methods 2016, 125, 19–27.
- Weerakoon, K.G.; Gordon, C.A.; Cai, P.; Gobert, G.N.; Duke, M.; Williams, G.M.; McManus, D.P. A novel duplex ddPCR assay for the diagnosis of schistosomiasis japonica: Proof of concept in an experimental mouse model. Parasitology 2017, 144, 1005–1015.
- Weerakoon, K.G.; Gordon, C.A.; Williams, G.M.; Cai, P.; Gobert, G.N.; Olveda, R.M.; Ross, A.G.; Olveda, D.U.; McManus, D.P. Droplet digital PCR diagnosis of human schistosomiasis: Parasite cell-free DNA detection in diverse clinical samples. J. Infect. Dis. 2017, 216, 1611–1622.
- Cai, P.; Weerakoon, K.G.; Mu, Y.; Olveda, R.M.; Ross, A.G.; Olveda, D.U.; McManus, D.P. Comparison of kato katz, antibody-based elisa and droplet digital PCR diagnosis of schistosomiasis japonica: Lessons learnt from a setting of low infection intensity. PLoS Negl. Trop. Dis. 2019, 13, e0007228.
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63.
- García-Bernalt, J.D.; Fernández-Soto, P.A.; Febrer-Sendra, B.; Crego-Vicente, B.; Muro, A.A. Loop-mediated isothermal amplification in schistosomiasis. J. Clin. Med. 2021, 10, 511.
- Avendaño, C.A.; Patarroyo, M.A. Loop-mediated isothermal amplification as point-of-care diagnosis for neglected parasitic infections. Int. J. Mol. Sci. 2020, 21, 7981.
- Tomita, N.; Mori, Y.; Fau-Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882.
- Hsieh, K.; Mage, P.L.; Csordas, A.T.; Eisenstein, M.; Soh, H.T. Simultaneous elimination of carryover contamination and detection of DNA with uracil-DNA-glycosylase-supplemented loop-mediated isothermal amplification (UDG-LAMP). Chem. Commun. 2014, 50, 3747–3749.
- Ma, C.; Wang, F.; Wang, X.; Han, L.; Jing, H.; Zhang, H.; Shi, C. A novel method to control carryover contamination in isothermal nucleic acid amplification. Chem. Commun. 2017, 53, 10696–10699.
- Deng, P.C.; Zhang, R.J.; Liang, Y.; Zhang, Y.K.; Yang, Q.L. Detection of Schistosoma japonicum—Infected Oncomelania hupensis by loop—Mediated isothermal amplification. Chin. J. Vet. Sci. 2009, 29, 1017–1018+1027. (In Chinese)
- Yang, Q.L.; Xu, L.F.; Zhang, Y.K.; Wang, K.G. Detection of Schistosoma japonicum cercaria DNA by loop-mediated isothermal amplification. Chin. J. Schistosomiasis Control 2008, 20, 209–211. (In Chinese)
- Tong, Q.B.; Chen, R.; Zhang, Y.; Yang, G.J.; Kumagai, T.; Furushima-Shimogawara, R.; Lou, D.; Yang, K.; Wen, L.Y.; Lu, S.H.; et al. A new surveillance and response tool: Risk map of infected Oncomelania hupensis detected by loop-mediated isothermal amplification (LAMP) from pooled samples. Acta Trop. 2015, 141, 170–177.
- Yu, C.X.; Yin, X.R.; Hua, W.Q.; Gao, Q. Establishment of loop mediated isothermal DNA amplification for identifying oncomelania snails infected with Schistosoma japonicum. J. Pathog. Biol. 2008, 3, 661–664. (In Chinese)
- Yu, C.X.; Yin, X.R.; Wang, J.; Song, L.J.; Qian, C.Y.; Wu, F.; He, W.; Zhang, W. Creation and use of a LAMP kit to rapidly detect snails infected with Schistosoma japonicum. J. Pathog. Biol. 2011, 6, 121–124. (In Chinese)
- Feng, J.T.; Xing, W.W.; Sun, K.; Yu, X.L.; Luo, Z.H.; Mao, J.W.; Xu, D.G. Application of visible loop-mediated isothermal amplification (LAMP) technologies in detecting oncomelania infected with Schistosoma japonicum. Mil. Med. Sci. 2016, 40, 133–136. (In Chinese)
- Xu, J.; Guan, Z.X.; Zhao, B.; Wang, Y.Y.; Cao, Y.; Zhang, H.Q.; Zhu, X.Q.; He, Y.K.; Xia, C.M. DNA detection of Schistosoma japonicum: Diagnostic validity of a lamp assay for low-intensity infection and effects of chemotherapy in humans. PLoS Negl. Trop. Dis. 2015, 9, e0003668.
- Wang, C.; Chen, L.; Yin, X.; Hua, W.; Hou, M.; Ji, M.; Yu, C.; Wu, G. Application of DNA-based diagnostics in detection of schistosomal DNA in early infection and after drug treatment. Parasites Vectors 2011, 4, 164.
- Wang, C.; Chuan-Xin, Y.U.; Min-Jun, J.I.; Song, L.J.; Yin, X.R.; Qian, C.Y.; Wang, J.; Guan-Ling, W.U. Use of loop-mediated isothermal amplification (LAMP) to detect whole blood samples infected with Schistosoma japonicum. J. Pathog. Biol. 2010, 5, 749–753. (In Chinese)
- Xiong, C.R.; Yin, X.R.; Song, L.J.; Shen, S.; Wang, J.; Gao, G.; Liu, Q.; Yu, C.X.; Yang, K. Comparison of the effectiveness of loop-mediated isothermal DNA amplification (LAMP) and mocroscopic dissection at detecting snails infected with Schistosoma japonicum. J. Pathog. Biol. 2014, 9, 1084–1087. (In Chinese)
- Li, Y.L.; Dang, H.; Guo, S.Y.; Cao, C.L.; Lü, S.; Xu, J.; Li, S.Z. National surveillance of Oncomelania hupensis in China. Chin. J. Schistosomiasis Control 2021, 33, 127–132. (In Chinese)
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204.
- Li, J.A.; Macdonald, J.A.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2018, 144, 31–67.
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Analyt. Chem. 2018, 98, 19–35.
- Wang, S.L.; Wang, L.P.; Wu, L.L.; Li, Y.L.; Zhang, L.J.; Lü, S.; Xu, J. Diagnostic value of nucleic acid detection in schistosomiasis japonica: A meta-analysis. Chin. J. Schistosomiasis Control 2020, 32, 15–22. (In Chinese)
- Song, Z.; Ting, L.; Kun, Y.; Wei, L.; Jian-Feng, Z.; Li-Chuan, G.; Yan-Hong, L.; Yang, D.; Qing-Jie, Y.; Hai-Tao, Y. Establishment of a recombinase-aided isothermal amplification technique to detect Schistosoma japonicum specific gene fragments. Chin. J. Schistosomiasis Control 2018, 30, 273–277. (In Chinese)
- Zhao, S.; Liu, Y.H.; Li, T.; Li, W.; Zhang, J.F.; Guo, L.C.; Ying, Q.J.; Yang, H.T.; Yang, K. Rapid detection of Schistosoma japonicum specific gene fragment by recombinase aided isothermal amplification combined with fluorescent probe. Chin. J. Parasitol. Parasit. Dis. 2019, 37, 23–27. (In Chinese)
- Ting, L.; Yan-Hong, L.; Song, Z.; Chun-Rong, X.; Xuan, D.; Jian-Feng, Z.; Wei, L.; Qing-Jie, Y.; Kun, Y. Rapid detection of Schistosoma japonicum-infected snails with recombinase-aided isothermal amplification assay. Chin. J. Schistosomiasis Control 2019, 31, 109–114. (In Chinese)
- Dong, X.; Xiong, C.R.; Li, T.; Zhao, S.; Zhang, J.F.; Li, W.; Yang, K. Early detection of Oncomelania snails infected eith Schistosoma japonicun by recombinase aided amplification. J. Pathog. Biol. 2019, 14, 1245–1249. (In Chinese)
- Ye, Y.Y.; Zhao, S.; Liu, Y.H.; Zhang, J.F.; Xiong, C.R.; Ying, Q.J.; Yang, K. Establishment of a nucleic acid dipstick test for detection of Schistosoma japonicum specific gene fragments based on the recombinase-aided isothermal amplification assay. Chin. J Schistosomiasis Control 2021, 33, 334–338. (In Chinese)
- Ye, Y.Y.; Zhao, S.; Liu, Y.H.; Bi, N.N.; Dong, X.; Xiong, C.R.; Zhu, H.R.; Tang, F.; Wang, X.Y.; Zhang, J.F.; et al. Performance of a recombinase-aided amplification assay for detection of Schistosoma japonicum infections in Oncomelania hupensis. Chin. J. Schistosomiasis Control 2021, 33, 185–188. (In Chinese)
- Sun, K.; Xing, W.; Yu, X.; Fu, W.; Wang, Y.; Zou, M.; Luo, Z.; Xu, D. Recombinase polymerase amplification combined with a lateral flow dipstick for rapid and visual detection of Schistosoma japonicum. Parasites Vectors 2016, 9, 476.
- Zhang, L.J.; Li, S.Z.; Wen, L.Y.; Lin, D.D.; Abe, E.M.; Zhu, R.; Du, Y.; Lv, S.; Xu, J.; Webster, B.L.; et al. The establishment and function of schistosomiasis surveillance system towards elimination in the People’s Republic of China. Adv. Parasitol. 2016, 92, 117–141.
- Xing, W.; Yu, X.; Feng, J.; Sun, K.; Fu, W.; Wang, Y.; Zou, M.; Xia, W.; Luo, Z.; He, H.; et al. Field evaluation of a recombinase polymerase amplification assay for the diagnosis of Schistosoma japonicum infection in hunan province of China. BMC Infect. Dis. 2017, 17, 164.
- Deng, W.P.; Xu, B.; Hong, Q.H.; Wang, S.L.; Lv, C.; Li, Y.L.; Song, S.P.; Chen, J.H.; Xu, J.; Li, S.Z. Establishment of the detection method for Schistosoma japonicum by recombinase polymerase amplification combined with electrochemical DNA biosensor. Chin. J. Parasitol. Parasit. Dis. 2020, 38, 42–48. (In Chinese)
- Wang, S.L.; Deng, W.P.; Li, Y.L.; Wang, L.P.; Zhang, L.J.; Lv, S.; Xu, J. Establishment of recombinase polymerase amplification technique for rapid detection of Schistosoma japonicum nucleic acid. Chin. J. Parasitol. Parasit. Dis. 2020, 38, 293–298. (In Chinese)
- Deng, W.P.; Hong, Q.H.; Xu, B.; Wang, S.L.; Wang, L.P.; Xu, J.; Hu, W.; Zhou, X.N. Development and preliminary evaluation of a rapid visualization detection method for circulating nucleic acids of Schistosoma japonicum based on RPA-LFD. Chin. J. Parasitol. Parasit. Dis. 2020, 38, 286–292. (In Chinese)
