Cytogenetics is the branch of genetics, cytology, and cell biology that analyses the nuclear genomes at the chromosome level. Cytogenetics makes the chromosome a substantial target in elementary plant cell biology and other fields such as mutagenesis and genotoxicity studies. Standard cytogenetic methods were, and are still, commonly used. Modern cytogenetic technologies involving advanced microscopy and imaging methods, that progress in the analyses on epigenetic DNA and histone modifications as well as DNA damage by using fluorescent antibodies benefit plant genome structure, dynamics, and evolution. They have also served the comprehensive evaluation of the effects of various mutagens on the plant genome that are observed as chromosome aberrations, including micronuclei (MN).
- cytogenetics
- micronuclei
- plant
1. The Importance of the Micronucleus Assay in Plants
2. Micronuclei—The Formation and Fate
3. Fluorescence In Situ Hybridization Serves to Understand the Origin of Micronuclei
References
- Evans, H.; Neary, G.; Williamson, F. The Relative Biological Efficiency of Single Doses of Fast Neutrons and Gamma-rays on Vicia Faba Roots and the Effect of Oxygen. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1959, 1, 216–229.
- Araldi, R.P.; de Melo, T.C.; Mendes, T.B.; de Sá Júnior, P.L.; Nozima, B.H.N.; Ito, E.T.; de Carvalho, R.F.; de Souza, E.B.; de Cassia Stocco, R. Using the comet and micronucleus assays for genotoxicity studies: A review. Biomed. Pharmacother. 2015, 72, 74–82.
- Mišík, A.; Nersesyan, A.; Mišíková, K.; Knasmueller, S. Micronucleus Assays with Meiotic Pollen Tetrad Cells of Tradescantia and with Mitotic Root Tip Cells of Allium cepa and Vicia faba Issues in Toxicology. In The Micronucleus Assay in Toxicology; Knasmüller, S., Fenech, M., Eds.; The Royal Society of Chemistry: London, UK, 2019.
- Chen, L.; Yuan, S.; Liu, X.; Zhou, X.; Zhou, Y.; Song, Y. Genotoxicity response of Vicia faba seedlings to cadmium in soils as characterized by direct soil exposure and micronucleus test. Ecotoxicology 2019, 29, 65–74.
- Oubane, M.; Khadra, A.; Ezzariai, A.; El Fels, L.; Kouisni, L.; Hafidi, M. Micronucleus assay based on Vicia faba roots as a tool to assess the performances of wastewater treatment systems. Environ. Technol. Innov. 2020, 19, 100903.
- Klein, P.; Chauvey, L.; Kallerhoff, J.; Pinelli, E.; Morard, M.; Silvestre, J. A Tool Derived from the Vicia faba Micronucleus Assay, to Assess Genotoxicity, Cytotoxicity or Biostimulation of Novel Compounds Used in Agriculture. Agronomy 2021, 11, 321.
- Ghosh, M.; Jana, A.; Sinha, S.; Jothiramajayam, M.; Nag, A.; Chakraborty, A.; Mukherjee, A.; Mukherjee, A. Effects of ZnO nanoparticles in plants: Cytotoxicity, genotoxicity, deregulation of antioxidant defenses, and cell-cycle arrest. Mutat. Res. Toxicol. Environ. Mutagen. 2016, 807, 25–32.
- Scherer, M.D.; Sposito, J.C.V.; Falco, W.F.; Grisolia, A.B.; Andrade, L.H.C.; Lima, S.M.; Machado, G.; Nascimento, V.A.; Gonçalves, D.A.; Wender, H.; et al. Cytotoxic and genotoxic effects of silver nanoparticles on meristematic cells of Allium cepa roots: A close analysis of particle size dependence. Sci. Total Environ. 2019, 660, 459–467.
- Gosh, M.; Ghosh, I.; Godderis, L.; Hoet, P.; Mukherjee, A. Genotoxicity of engineered nanoparticles in higher plants. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2019, 842, 132–145.
- Prabhu, S.; Kumar, V.; Bharath, J.; Thaha, M.; Rajeswari, D. A Review on Vicia faba as a plant test system in toxicity evaluation of various metals: Vicia-micronucleus test (Vicia-MCN). Int. J. Chem. Tech. Res. 2017, 10, 961–967.
- Jiang, X.; Chen, H.; Liao, Y.; Ye, Z.; Li, M.; Klobu, G. Ecotoxicity and genotoxicity of polystyrene microplastics on higher plant Vicia faba. Environ. Pollut. 2019, 251, 831–838.
- Kaur, M.; Sharma, A.; Soodan, R.K.; Chahal, V.; Kumar, V.; Katnoria, J.K.; Nagpal, A.K. Allium cepa Root Chromosomal Aberration Assay: A Tool to Assess Genotoxicity of Environmental Contaminants. In Environmental Contaminants and Natural Products: A Human Health Perspective; Sharma, A., Kumar, M., Kaur, S., Nagpal, A.K., Eds.; Bentham Science: Sharjah, United Arab Emirates, 2019; pp. 65–93.
- Zgórska, A.; Borgulat, A. Genotoxicity of wastewater samples from the textile industry detected by broad bean (Vicia faba) micronucleus test assay. Appl. Ecol. Environ. Res. 2020, 18, 5315–5323.
- Abdelsalam, N.R.; Abdel-Megeed, A.; Alic, H.M.; Saleme, M.Z.M.; Al-Hayalif, M.F.A.; Elshikh, M.S. Genotoxicity effects of silver nanoparticles on wheat (Triticum aestivum L.) root tip cells. Ecotoxicol. Environ. Saf. 2018, 155, 76–85.
- Alvarenga, I.F.S.; dos Santos, F.E.; Silveira, G.L.; Andrade-Vieira, L.F.; Martin, G.C.; Guilherme, L.R.G. Investigating arsenic toxicity in tropical soils: A cell cycle and DNA fragmentation approach. Sci. Total Environ. 2020, 698, 134272.
- Korshikov, I.I.; Belonozhko, Y.A.; Lapteva, E.V. The Use of a Micronucleus Test in Pinus pallasiana D. Don and Picea abies (L.) Karst. for the Assessment of Technogenic Pollution’s Influence. Cytol. Genet. 2019, 53, 106–112.
- Mishra, K. Assessment of Cytotoxic and Genotoxic Potential of Heavy Metals in Plants: A Review. Int. J. Plant Environ. 2020, 6, 152–155.
- Heddle, J.A.; Fenech, M.; Hayashi, M.; MacGregor, J.T. Reflections on the development of micronucleus assays. Mutagenesis 2010, 26, 3–10.
- Luzhna, L.; Kathiria, P.; Kovalchuk, O. Micronuclei in genotoxicity assessment: From genetics to epigenetics and beyond. Front. Genet. 2013, 4, 131.
- Kwon, M.; Leibowitz, M.L.; Lee, J.H. Small but mighty: The causes and consequences of micronucleus rupture. Exp. Mol. Med. 2020, 52, 1777–1786.
- Nersesyan, A.; Fenech, M.; Bolognesi, C.; Misik, M.; Setayesh, T.; Wultsch, G.; Bonass, S.; Thomas, P.; Knasmuller, S. Use of the lymphocyte cytokinesis-block micronucleus assay in occupational biomonitoring of genome damage caused by in vivo exposure to chemical genotoxins: Past, present and future. Mutat. Res. 2016, 770, 1–11.
- McClelland, S.E. Role of chromosomal instability in cancer progression. Endocr.-Relat. Cancer 2017, 24, T23–T31.
- Saks, M.; Upreti, S.; Rajendra, S.V.; Dang, R. Genotoxicity: Mechanisms, testing guidelines and methods. Glob. J. Pharm. Pharm. Sci. 2017, 1, 133–138.
- Kirsch-Volders, M.; Fenech, M.; Bolognesi, C. Validity of the lymphocyte cytokinesis-block micronucleus assay (L-CBMN) as biomarker for human exposure to chemicals with different modes of action: A synthesis of systematic reviews. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018, 836, 47–52.
- Ye, C.J.; Sharpe, Z.; Alemara, S.; MacKenzie, S.; Liu, G.; Abdallah, B.; Horne, S.; Regan, S.; Heng, H.H. Micronuclei and Genome Chaos: Changing the System Inheritance. Genes 2019, 10, 366.
- Norppa, H.; Falck, G.C.M. What do human micronuclei contain? Mutagenesis 2003, 18, 221–233.
- Sommer, S.; Buraczewska, I.; Kruszewski, M. Micronucleus Assay: The State of Art, and Future Directions. Int. J. Mol. Sci. 2020, 21, 1534.
- Fenech, M. Cytokinesis-block micronucleus cytome assay evolution into a more comprehensive method to measure chromosomal instability. Genes 2020, 11, 1203.
- Hayashi, M. The micronucleus test—Most widely used in vivo genotoxicity test. Genes Environ. 2016, 38, 18.
- Hintzsche, H.; Hemmann, U.; Poth, A.; Utesch, D.; Lott, J.; Stopper, H. Fate of micronuclei and micronucleated cells. Mutat. Res. 2017, 771, 85–98.
- Liehr, T. Fluorescence In Situ Hybridization (FISH)—Application Guide, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2017.
- Liehr, T. Is molecular cytogenetic diagnostic of are diseases in Europe close to extinction. J. Genet. Genom. 2020, 4, 2.
- Florian, R.T.; Kraft, F.; Leitão, E.; Kaya, S.; Klebe, S.; Magnin, E.; van Rootselaar, A.-F.; Buratti, J.; Kühnel, T.; Schröder, C.; et al. Unstable TTTA/TTTCA expansions in MARCH6 are associated with familial adult myoclonic epilepsy type 3. Nat. Commun. 2019, 10, 4919.
- Weiss, H.; Maluszynska, J. Chromosomal rearrangement in autotetraploid plants of Arabidopsis thaliana. Hereditas 2000, 133, 255–261.
- Siroky, J.; Zluvova, J.; Riha, K.; Shippen, R.E.; Vyskot, B. Rearrangements of ribosomal DNA clusters in late generation telomerase-deficient Arabidopsis. Chromosoma 2003, 112, 116–123.
- Juchimiuk, J.; Hering, B.; Maluszynska, J. Multicolour FISH in an analysis of chromosome aberrations induced by N-nitroso-N-methylurea and maleic hydrazide in barley cells. J. Appl. Genet. 2007, 48, 99–106.
- Juchimiuk-Kwasniewska, J.; Brodziak, L.; Maluszynska, J. FISH in analysis of gamma ray-induced micronuclei formation in barley. J. Appl. Genet. 2010, 52, 23–29.
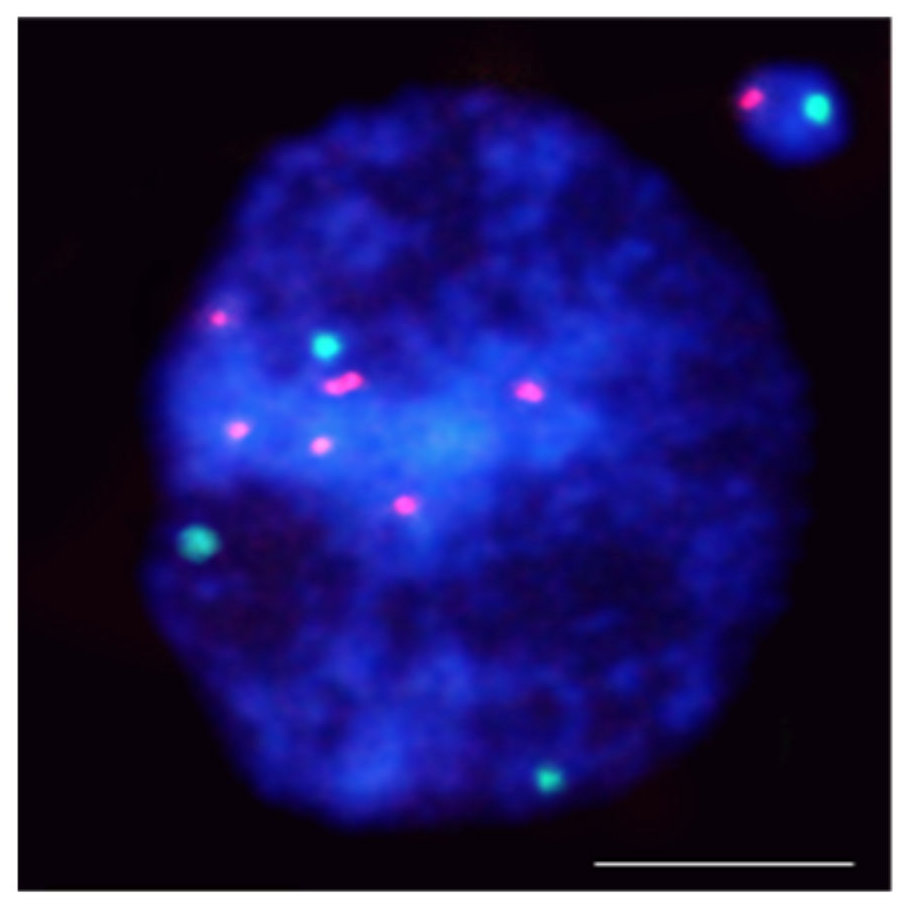
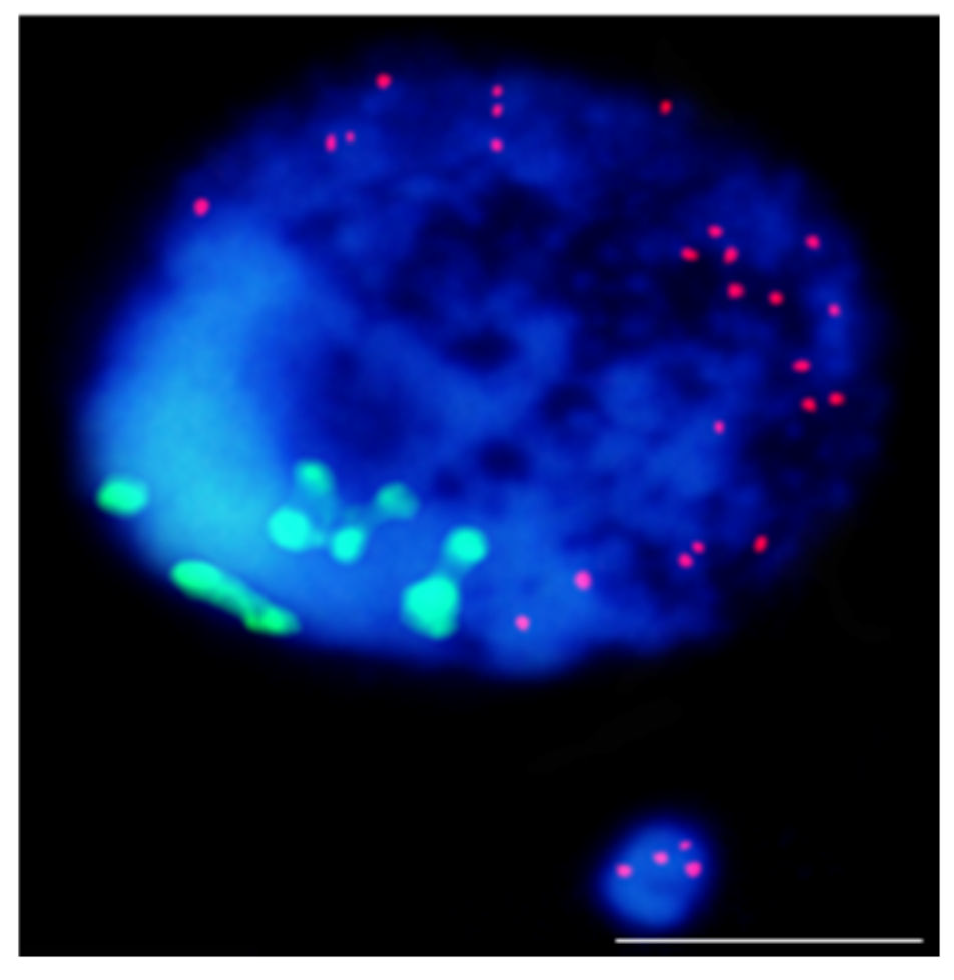
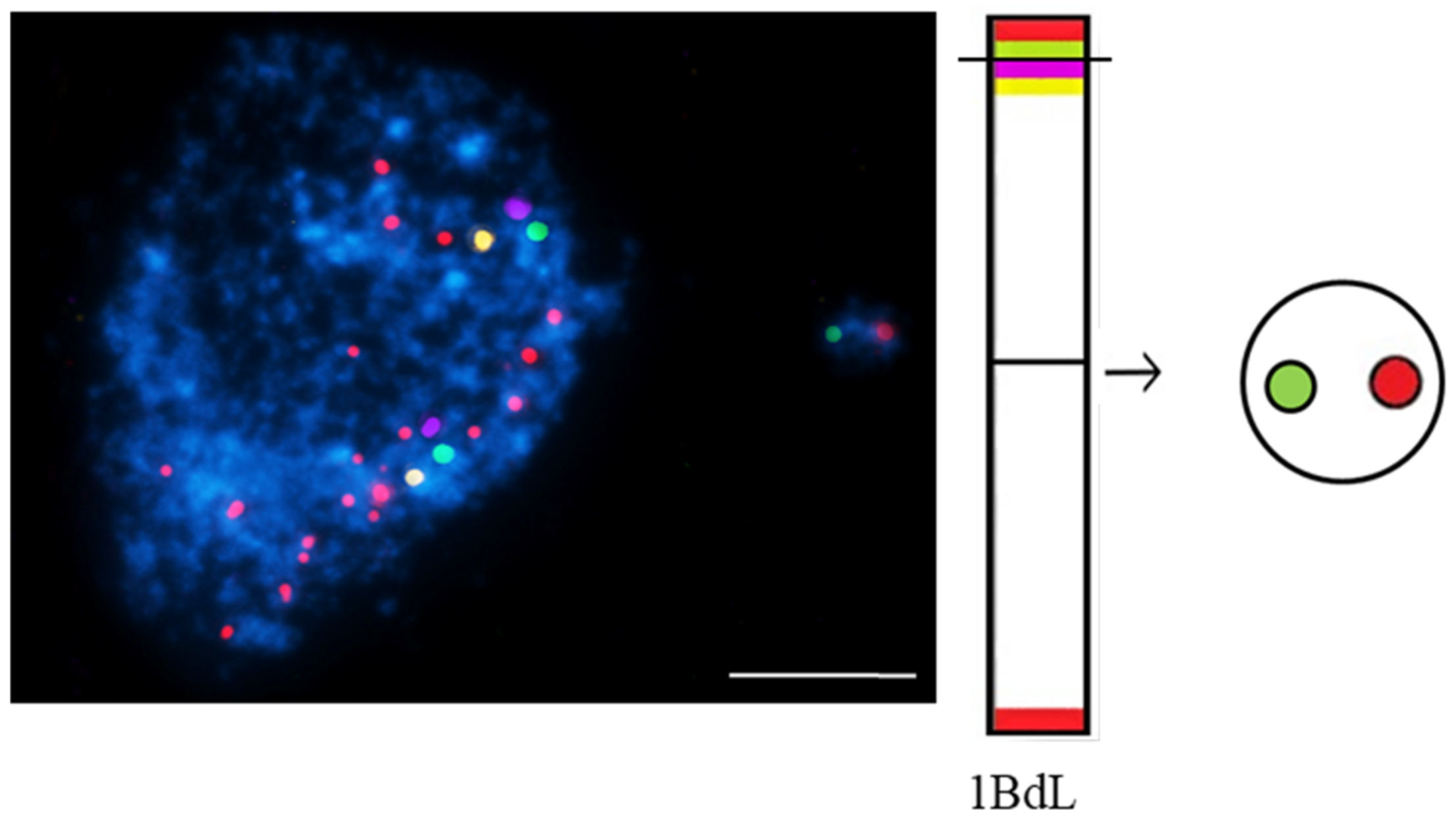
- Kus, A.; Kwasniewska, J.; Hasterok, R. Brachypodium distachyon—A Useful Model in the Qualification of Mutagen-Induced Micronuclei Using Multicolor FISH. PLoS ONE 2017, 12, e0170618.
- Rocha, L.C.; Mittelmann, A.; Houben, A.; Techio, V.H. Fragile sites of 45S rDNA of Lolium multiflorum are not hotspots for chromosomal breakages induced by X-ray. Mol. Biol. Rep. 2016, 43, 659–665.
- Hasterok, R.; Jenkins, G.; Langdon, T.; Jones, R.N. The nature and destiny of translocated B-chromosome-specific satellite DNA of rye. Chromosome Res. 2002, 10, 83–86.
- Hasterok, R.; Langdon, T.; Taylor, S.; Jenkins, G. Combinatorial labeling of DNA probes enables multicolor fluorescence in situ hybridization in plants. Folia Histochem. Cytobiol. 2002, 40, 319–323.
- Nederlof, P.M.; Robinson, D.; Abuknesha, R.; Wiegant, J.; Hopman, A.H.N.; Tanke, H.J.; Raap, A.K. Three-color fluorescence in situ hybridization for the simultaneous detection of multiple nucleic acid sequences. Cytom. J. Int. Soc. Anal. Cytol. 1989, 10, 20–27.
- Natarajan, A.; Boei, J. Formation of chromosome aberrations: Insights from FISH. Mutat. Res. Mol. Mech. Mutagen. 2003, 544, 299–304.
- Lusinska, J.; Majk, J.; Betekhtin, A.; Susek, K.; Wolny, E.; Hasterok, R. Chromosome identification and reconstruction of evolutionary rearrangements in Brachypodium distachyon, B. stacei and B. hybridum. Ann. Bot. 2018, 122, 445–459.
- He, L.; Braz, G.T.; Torres, G.; Jiang, J. Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species. Chromosoma 2018, 127, 505–513.
- Hou, L.; Xu, M.; Zhang, T.; Xu, Z.; Wang, W.; Zhang, J.; Yu, M.; Ji, W.; Zhu, C.; Gong, Z.; et al. Chromosome painting and its applications in cultivated and wild rice. BMC Plant Biol. 2018, 18, 110.
- Chen, F.; Song, Y.; Li, X.; Chen, J.; Mo, L.; Zhang, X.; Lin, Z.; Zhang, L. Genome sequences of horticultural plants: Past, present, and future. Hortic. Res. 2019, 6, 112.
- Albert, P.S.; Zhang, T.; Semrau, K.; Rouillard, J.-M.; Kao, Y.-H.; Wang, C.-J.R.; Danilova, T.V.; Jiang, J.; Birchler, J.A. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. USA 2019, 116, 1679–1685.
- Meng, Z.; Wang, Q.; Khurshid, H.; Raza, G.; Han, J.; Wang, B.; Wang, K. Chromosome Painting Provides Insights Into the Genome Structure and Evolution of Sugarcane. Front. Plant Sci. 2021, 12, 731664.
- Hovhannisyan, G.; Aroutiounian, R.; Liehr, T. Chromosomal Composition of Micronuclei in Human Leukocytes Exposed to Mitomycin C. J. Histochem. Cytochem. 2012, 60, 316–322.
- Lysak, M.A.; Berr, A.; Pecinka, A.; Schmidt, R.; McBreen, K.; Schubert, I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 2006, 103, 5224–5229.
- Idziak, D.; Hazuka, I.; Poliwczak, B.; Wiszynska, A.; Wolny, E.; Hasterok, R. Insight into the Karyotype Evolution of Brachypodium Species Using Comparative Chromosome Barcoding. PLoS ONE 2014, 9, e93503.
- Kus, A.; Kwasniewska, J.; Szymanowska-Pulka, J.; Hasterok, R. Dissecting the chromosomal composition of mutagen-induced micronuclei in Brachypodium distachyon using multicolour FISH. Ann. Bot. 2018, 122, 1161–1171.
- Kus, A.; Szymanowska-Pułka, J.; Kwasniewska, J.; Hasterok, R. Detecting Brachypodium distachyon Chromosomes Bd4 and Bd5 in MH- and X-Ray-Induced Micronuclei Using mcFISH. Int. J. Mol. Sci. 2019, 20, 2848.
