More than 90% of all head and neck cancers (HNCs) are head and neck squamous cell carcinomas (HNSCCs) arising from the mucosal surfaces of the upper aerodigestive tract. HNSCCs are the the sixth most prevalent cancer worldwide, and are often associated with either carcinogens, such as alcohol and tobacco use, or oncogenic human papillomavirus (HPV) infection. HNSCCs have been found to be diverse with a high rate of genetic heterogeneity, resulting in hyper-activation of oncogenes (e.g.,
PIK3CA
and
HRAS
) and loss-of-function mutations in tumor suppressor genes (e.g.,
TP53
,
CASP8
, and
NOTCH1
). HNSCC cohorts from The Cancer Genome Atlas (TCGA) RNA-seq data and clinical data show patients with
PIK3CA
alterations, including amplification and gain, also have a higher chance of harboring
TP53
mutations. In addition, these patients bearing both mutations have a significantly worse 10-year survival prognosis compared with their wildtype cohort counterparts.
- HNSCC translational research
- HNSCC preclinical models
- Head and neck squamous cell carcinoma
- PIK3CA/TP53
- TCGA
- gene mutations
- deletions
- amplifications
- Squamous Cell Carcinoma
- SCC
1. Introduction
Head and neck cancers (HNC) are a heterogeneous group of tumors arising from the mucosal surfaces of the upper aerodigestive tract [1]. Collectively, HNC is the sixth most prevalent cancer worldwide [1]. Some 90% of all HNCs are head and neck squamous cell carcinomas (HNSCCs) and HNSCCs are often associated with either carcinogens, such as alcohol and tobacco use, or oncogenic human papillomavirus (HPV) infection [2][3][2,3], thereby categorized as HPV(−) or HPV(+) HNSCCs. HNSCCs have been found to be diverse with a high rate of genetic heterogeneity, resulting in hyper-activation of oncogenes (e.g., PIK3CA and HRAS) and loss-of-function mutations in tumor suppressor genes (e.g., TP53, CASP8, and NOTCH1) [4][5][4,5]. Phosphoinositide 3-kinase (PI3K) is a frequently deregulated pathway in HNSCCs with a phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) gene mutation rate of approximately 16% and gene amplification of more than 30% in tumors [6][7][6,7]. PI3Ks are activated by receptor tyrosine kinases (RTKs), such as epidermal growth factor receptor (EGFR), and consist of different classes of enzymes vital for differentiation, proliferation and cell survival [8]. Mammalian target of rapamycin (mTOR) complexes (mTORC1 and mTORC2) and protein kinase B (also known as AKT) are also involved in this pathway that can activate transcription and other signaling molecules of the PI3K pathway [9]. Monoclonal antibodies (mAbs) that inhibit EGFR have been used for both HPV(−) and HPV(+) subtypes of HNSCCs; however, they were found to have limited efficacy and elicited resistance [10].
Another highly mutated gene in HNSSCs is the tumor protein p53 (TP53) gene, with over 80% of HPV(−) HNSCCs harboring loss-of-function mutations in TP53; however, TP53 mutations occur much less frequently in HPV(+) HNSCCs (~3%) [4]. TP53 is a tumor-suppressor gene encoding a transcription factor that maintains DNA repair, cell cycle, senescence and apoptosis [11]. These attributes make p53 an important cell sensor for oncogene activation and DNA damage. It has been found that the degradation of p53 is associated with HPV E6 oncoproteins [3]. Although there have been several therapies that target p53 in hopes to restore p53 function, they have yet to be proven effective in clinical trials [12]. By and large, TP53 mutations are associated with poor HNSCC prognosis and overall survival with increased rate of recurrence and resistance to therapies. It remains poorly understood whether HNSCCs harboring different genetic alterations exhibit differential immune tumor microenvironment (TME). For instance, it is unknown whether HNSCCs with the double mutations in TP53 and PIK3CA have a more immunosuppressive TME.
Prior studies have generated murine models that mimicked the alterations of PIK3CA or p53 in HNSCCs. Transgenic mice that overexpressed wild-type PIK3CA in head and neck epithelium were generated; however, PIK3CA overexpression alone was not sufficient to initiate HNSCC formation [6]. Nevertheless, these PIK3CA Tg mice were much more susceptible to 4-nitroquinoline 1 oxide (4NQO)-induced HNSCC carcinogenesis [6]. Conditional deletion of p53 in mouse epithelial cells with K14.CrePR1 led to SCC development in about half of mice after 20 months [13]. To establish a mouse model that more closely resembles the genetic alterations in HNSCCs and allows us to better investigate immune evasion mechanisms of HNSCCs, we generated a novel genetic model by deleting p53 and constitutively activating PIK3CA in mouse keratin 15-expressing (K15+) stem cells, which leads to the development of multilineage tumors including SCCs, termed Keratin-15-p53-PIK3CA (KPPA) tumors.
2. HNSCC Patients with Double Genetic Alterations in PIK3CA and TP53 Exhibited Worse Prognosis and More Immunosuppressive TME
The Cancer Genome Atlas (TCGA) RNA-seq data and clinical data of HNSCC cohorts were obtained from the cBioPortal (hhttps://cbioportal.org) (see dettp://cbails ioportal.orgn Methods). In analyzing the dataset of HNSCC patients (TCGA, PanCancer Atlas, n = 489 samples), we found that the patients with PIK3CA alterations, including amplification and gain, also have a higher chance of harboring TP53 mutations (Figure 1A). After merging two datasets (see details in Methods), we divided the patients into four different groups: PIK3CAAmp/TP53Mutated (n = 294), PIK3CAAmp/TP53WT (n = 85), PIK3CAWT/TP53Mutated (n = 56), and PIK3CAWT/TP53WT (n = 54). Kaplan Meier curves of five-year survival were shown for four different groups, and PIK3CAAmp/TP53Mutated group exhibited a hazard ratio of 1.61 (95% CI, 0.94–2.75) compared to PIK3CAWT/TP53WT group (Figure 1B). PIK3CAAmp/TP53Mutated group’s 10-year survival hazard ratio was 1.8 (95% CI, 1.06–3.07), and showed a significantly worse prognosis than PIK3CAWT/TP53WT group (Supplemental Figure S1).
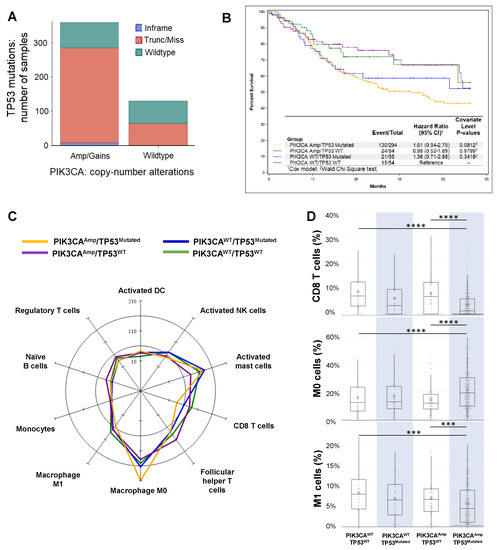
Figure 1. Analysis of The Cancer Genome Atlas (TCGA) datasets of Head and Neck Squamous cell carcinoma (HNSCC) patients. (A) Association between PIK3CA gene alterations and TP53 gene mutation in HNSCC patients (TCGA, PanCancer Atlas, n = 489 samples). The Cancer Genome Atlas (TCGA) RNA-seq data and clinical data of HNSCC cohorts were obtained from the cBioPortal (hhttps://cbioportal.org) (see dettp://cbails ioportal.orgn Method). Patients with PIK3CAAmp/gain (n = 359) had a higher chance to harbor TP53 mutations than patients with PIK3CAWT (n = 130). (B) Kaplan-Meier overall 5-year survival curves of HNSCC patients in 4 groups (PIK3CAAmp/TP53Mutated, PIK3CAAmp/TP53WT, PIK3CAWT/TP53Mutated, and PIK3CAWT/TP53WT) as described in Methods. Only patients with available survival data were included for this analysis (n = 487). (C) A radar plot of the cell types that reached significance in the omnibus Kruskal-Wallis test when comparing among the 4 groups. The scale is per 1000 cells. (D) Box and whisker plots of CD8 T cells, resting macrophages (M0), and M1 macrophages. The expression of CD8 T cell signature genes: PIK3CAAmp/TP53Mutated group (3.77 ± 5.32) was significantly lower than groups of PIK3CAWT/TP53WT (8.97 ± 7.86) and PIK3CAAmp/TP53WT (8.22 ± 8.12). The expression of M0 signature genes: PIK3CAAmp/TP53Mutated group (21.07 ± 14.08) was significantly higher than groups of PIK3CAWT/TP53WT (15.25 ± 12.32) and PIK3CAAmp/TP53WT (13.93 ± 11.23). The expression of M1 signature genes: PIK3CAAmp/TP53Mutated group (5.35 ± 5.05) was significantly lower than groups of PIK3CAWT/TP53WT (8.17 ± 5.62) and PIK3CAAmp/TP53WT (6.92 ± 5.17).
We uploaded RNA-seq data of HNSCC patients onto CIBERSORT (see details in Methods), which estimated the relative proportions of 22 immune cell types (Supplemental Figure S2), with a more in-depth dissection shown in Supplemental Table S1. Both innate and adaptive immune cells varied in their expression levels depending on the genetic alterations in 4 groups (Figure 1C). In particular, we found that the expression of CD8 T cell signature genes was significantly lower in PIK3CAAmp/TP53Mutated group, compared with PIK3CAWT/TP53WT and PIK3CAAmp/TP53WT groups (Figure 1D). In addition, PIK3CAAmp/TP53Mutated group had significantly lower expression of activated NK cell-associated genes compared with PIK3CAWT/TP53WT and PIK3CAWT/TP53Mutated groups (Supplemental Table S1). PIK3CAAmp/TP53Mutated group expressed significantly higher level of resting macrophage (M0) signature genes but lower level of activated macrophage (M1) genes than PIK3CAWT/TP53WT and PIK3CAAmp/TP53WT groups (Figure 1D). We conclude that HNSCCs with the genotype of PIK3CAAmp/TP53Mutated appear to have a highly immunosuppressive TME.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Marsit, C.J.; McClean, M.D.; Nelson, H.H.; Christensen, B.C.; Haddad, R.I.; Clark, J.R.; Wein, R.O.; Grillone, G.A.; Houseman, E.A.; et al. Biomarkers of HPV in head and neck squamous cell carcinoma. Cancer Res. 2012, 72, 5004–5013. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xia, R.H.; Ye, D.X.; Li, J. Human Papillomavirus 16 Infection and TP53 Mutation: Two Distinct Pathogeneses for Oropharyngeal Squamous Cell Carcinoma in an Eastern Chinese Population. PLoS ONE 2016, 11, e0164491. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Perez Sayans, M.; Chamorro Petronacci, C.M.; Lorenzo Pouso, A.I.; Padin Iruegas, E.; Blanco Carrion, A.; Suarez Penaranda, J.M.; Garcia Garcia, A. Comprehensive Genomic Review of TCGA Head and Neck Squamous Cell Carcinomas (HNSCC). J. Clin. Med. 2019, 8, 1896. [Google Scholar] [CrossRef]
- Du, L.; Chen, X.; Cao, Y.; Lu, L.; Zhang, F.; Bornstein, S.; Li, Y.; Owens, P.; Malkoski, S.; Said, S.; et al. Overexpression of PIK3CA in murine head and neck epithelium drives tumor invasion and metastasis through PDK1 and enhanced TGFbeta signaling. Oncogene 2016, 35, 4641–4652. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Kang, H.; Mehra, R. Targeting phosphoinositide 3-kinase (PI3K) in head and neck squamous cell carcinoma (HNSCC). Cancers Head Neck 2018, 3, 3. [Google Scholar] [CrossRef]
- Du, L.; Shen, J.; Weems, A.; Lu, S.L. Role of phosphatidylinositol-3-kinase pathway in head and neck squamous cell carcinoma. J. Oncol. 2012, 2012, 450179. [Google Scholar] [CrossRef]
- Tan, B.S.; Tiong, K.H.; Choo, H.L.; Chung, F.F.; Hii, L.W.; Tan, S.H.; Yap, I.K.; Pani, S.; Khor, N.T.; Wong, S.F.; et al. Mutant p53-R273H mediates cancer cell survival and anoikis resistance through AKT-dependent suppression of BCL2-modifying factor (BMF). Cell Death Dis. 2015, 6, e1826. [Google Scholar] [CrossRef]
- De Pauw, I.; Lardon, F.; Van den Bossche, J.; Baysal, H.; Fransen, E.; Deschoolmeester, V.; Pauwels, P.; Peeters, M.; Vermorken, J.B.; Wouters, A. Simultaneous targeting of EGFR, HER2, and HER4 by afatinib overcomes intrinsic and acquired cetuximab resistance in head and neck squamous cell carcinoma cell lines. Mol. Oncol. 2018, 12, 830–854. [Google Scholar] [CrossRef]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, M.R.; Pan, Q. Novel p53 therapies for head and neck cancer. World J. Otorhinolaryngol. Head Neck Surg. 2016, 2, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Savar, A.; Acin, S.; Gonzalez, C.L.; El-Sawy, T.; Mejia, O.; Li, Z.; Esmaeli, B.; Lacy-Hulbert, A.; El-Naggar, A.K.; McCarty, J.H.; et al. Loss of epithelial p53 and αv integrin cooperate through Akt to induce squamous cell carcinoma yet prevent remodeling of the tumor microenvironment. Oncogene 2015, 34, 516–524. [Google Scholar] [CrossRef] [PubMed]
