You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 1 by Elizabeth Vincan and Version 2 by Peter Tang.
Organoids are defined as three-dimensional structures grown from stem cells and consist of organ-specific cell types that self-organize to recapitulate key features and functional characteristics of tissues in a dish. Organoids can be initiated from two main types of stem cells: (1) pluripotent embryonic stem (ES) cells or synthetically induced pluripotent stem (iPS) cells and (2) organ-restricted stem cells.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), which was classified as a pandemic in March 2020. Organoids from diverse human organs have been adopted to understand COVID-19 and the effects the virus has on organs.
- COVID-19
- SARS-CoV-2
- organoid
- ex vivo models
- infectious disease
1. Modelling Human Infectious Disease with Organoids
Organoids are defined as three-dimensional structures grown from stem cells and consist of organ-specific cell types that self-organize to recapitulate key features and functional characteristics of tissues in a dish [1]. Organoids can be initiated from two main types of stem cells: (1) pluripotent embryonic stem (ES) cells or synthetically induced pluripotent stem (iPS) cells and (2) organ-restricted stem cells. Pluripotent stem cell generated organoids were made more accessible by the discovery by Takahashi and Yamanaka that somatic cells could be taken back (“induced”) to pluripotency [2]. Organoids generated from organ-restricted stem cells were first described by the Clevers laboratory in 2009 [3] using mouse intestinal epithelium. Using variations of this intestinal epithelium protocol, organoids have since been established from many organs, especially epithelial tissues. A PubMed search on the term “organoid” shows an exponential increase in publications since 2009 (Figure 1a). Earlier use of the term in the period of 1965–1985 was initiated by developmental biologists to describe tissue explants. Organ-restricted stem cell-derived organoid technology was embraced to understand tissue homeostasis and regeneration and how cancers start in these tissues (reviewed in [1]). Once it was realized that patient tumor-derived organoids could also be established [4], this led to the establishment of tumor biobanks to understand the processes that lead to cancer growth and to guide treatment [5][6][7][5,6,7]. More recently, organoids are being adopted to study human infectious diseases (Figure 1b,c)
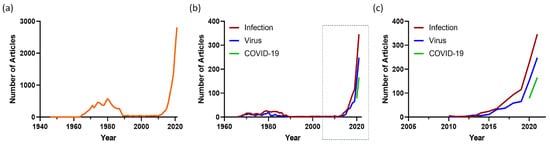
Figure 1. Organoid articles in PubMed. The number of articles published each year for the search term (a) “Organoid” and (b) “Organoid AND Infection” (red), “Organoid AND Virus” (blue) and “Organoid” AND COVID-19” (green) in PubMed. The boxed section of (b) is shown in (c).
2. Overview of SARS-CoV-2
2.1. Human Coronaviruses
Coronaviruses (CoVs) are enveloped, non-segmented, positive-sense single stranded RNA (ssRNA) viruses that belong to the Coronaviridea family and can infect humans and animals [8][16]. Seven CoVs are known to infect humans, four (human (H)CoV-NL63, HCoV-OC43, HCoV-229E and HCoV-HKU) of which cause mild, seasonal respiratory tract diseases. Three of the seven have emerged over the last 20 years causing severe human disease. Severe acute respiratory syndrome CoV (SARS-CoV), a beta coronavirus, emerged in late 2002 in Guangdong Province, China and spread rapidly from a single index case to five countries and led to large outbreaks, many in tertiary hospitals, with a case fatality of 9.6%. The virus spread to a further seven countries and, in all, resulted in 8098 cases globally with 774 deaths, including many healthcare workers [9][17]. SARS-CoV is thought to have originated from bats and transmitted from bats to wild animals, such as civets, and then to humans. The epidemic ended in 2004, despite human–human transmission, but possibly resulting from the relatively low infectivity of the virus. In 2012, Middle Eastern Respiratory Syndrome CoV (MERS-CoV) emerged, causing outbreaks of severe pneumonia in Saudi Arabia and South Korea that were associated with a high fatality rate of 34% [10][11][18,19]. Imported cases have been reported in several countries, including the United Kingdom, the US, Germany, Italy, Greece, The Netherlands, Malaysia and South Korea. The largest outbreak occurred in 2014 in Saudi Arabia, with over 500 hospitals involved within a few months. In 2015, a large outbreak across 16 hospitals arising from a single index case occurred in South Korea. The outbreak was finally stopped by quarantining over 17,000 individuals [10][18]. MERS-CoV is also thought to have originated in bats.
These two highly pathogenic HCoV and the large pool of SARS-CoV-like viruses discovered in bats [12][20] meant that further zoonotic CoV epidemics and pandemics were highly likely.
2.2. The Emergence of SARS-CoV-2
In December 2019, a novel CoV (initially named “2019-nCoV”) emerged in Wuhan, Hubei Province, China, causing severe pneumonia [13][21]. The virus was found to share 80% homology with SARS-CoV and was subsequently renamed as SARS-CoV-2. Genomic sequencing suggested that the reservoir for SARS-CoV-2 is likely to be bats and pangolins [14][15][16][22,23,24]. The disease caused by SARS-CoV-2 was soon named coronavirus disease 2019 (COVID-19) and declared a fast-evolving pandemic by the World Health Organization (WHO) on 11 March 2020. This triggered an unprecedented global effort to investigate the characteristics of the virus to develop strategies to diagnose, prevent and treat COVID-19. As of 22 January 2022, more than 347 million cases of COVID-19 have been diagnosed worldwide, with 5.6 million deaths, making it the deadliest pandemic since the influenza pandemic of 1918.
2.3. SARS-CoV-2 Cell Entry
SARS-CoV-2 particles comprise four main structural proteins: spike (S), envelope (E), membrane (M) and nucleocapsid (N) (Figure 2). CoV S proteins are composed of two domains, the receptor binding (S1) and fusion (S2) domains. Viral entry is initiated by the docking of the S protein [17][18][25,26] via its receptor binding domain (RBD) in S1 to its cellular receptor. Like SARS-CoV, SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) as its cellular receptor [19][20][27,28]. In the respiratory tract, the ACE2 receptor is preferentially expressed on the apical surface of epithelial cells. Both SARS-CoV and SARS-CoV-2 infect by binding apically distributed ACE2 and during their egress from infected cells, are also released from the apical surface of respiratory cells [21][22][23][24][29,30,31,32]. This is significant as the understanding of viral entry, release and neutralization may be guided very differently by studying these events in polarized, compared to non-polarized, respiratory epithelial cells.
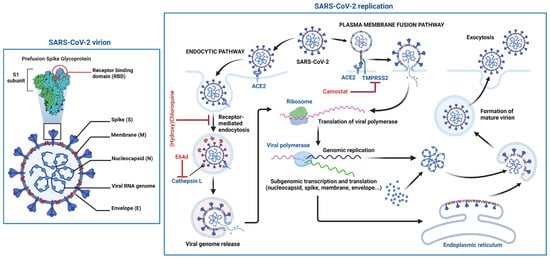

Figure 2. Schematic structure of the SARS-CoV-2 virion and the viral life cycle. The SARS-CoV-2 virion contains an envelope with three proteins: the spike (S) protein, membrane (M) and envelope (E) proteins. The viral RNA genome and the nucleocapsid (N) protein are contained inside the particle. The virion schematic shows in more detail the S protein subunit 1 (S1) containing the receptor binding domain (RBD). The SARS-CoV-2 replication cycle shows two modes of viral entry (endocytic and TMPRSS2-mediated pathways). Both require docking to ACE2 at the cell surface and release of the viral genome into the cytoplasm via membrane fusion. The endocytic pathway is inhibited by (hydroxy)chloroquine and E64d, while TMPRSS2-mediated entry is inhibited by camostat. After synthesis of viral genome and proteins, viral particles are assembled and exit the infected cell. Figure 2 was created with BioRender.com.
2.4. Emergence of SARS-CoV-2 Mutants
Since the emergence and persistence of SARS-CoV-2 in the global population, the virus has undergone several mutational shifts towards variants carrying combinations of changes within the S1 domain of the S protein, in addition to those already occurring in the RBD, enhancing their affinity and binding to the human ACE2 receptor. These mutations resulted in greater virulence and transmissibility and are potentially able to evade neutralizing antibodies (NAb) targeting the RBD and receptor binding motif (RBM) of the spike protein [25][26][27][28][38,39,40,41]. The most frequent variants have included Alpha (B.1.1.7) [29][42], Beta (B.1.351) [30][43], Delta (B.1.617.2) [31][44] and Epsilon (B.1.427/B.1.429) [32][45], which have been associated with greater transmission and disease severity and, for B.1.351 and B.1.427/B.1.429, escape from neutralization by vaccine-induced antibodies [32][33][34][45,46,47]. The most recent variant to emerge is Omicron (B.1.529) [35][36][48,49]. Being substantially more infectious than Delta, Omicron is not effectively inhibited by vaccine-induced neutralizing antibodies and, in individuals who have received only two doses of COVID-19 vaccines, is associated with reduced vaccine efficacy [37][38][39][40][50,51,52,53].
3. COVID-19 Is a Systemic Disease
SARS-CoV-2 is mainly transmitted through aerosolised respiratory droplets [41][54]. However, transmission by direct contact and through mucosal membranes is also possible, although they are less likely to transmit infection than close contact with an infected person [42][43][55,56]. The basic reproductive number (R0) of the ancestral Wuhan virus and early variants ranged from 2 to 4 [44][45][57,58]. For the Delta variant, the R0 is 97% higher than previous variants, making this virus substantially more transmissible [46][59]. However, with an R0 that is 4-fold higher than Delta, Omicron became the most transmissible SARS-CoV-2 variant to date [47][60].
The median incubation period for people who become symptomatic after contact with an infected person is 5 to 6 days for the Wuhan strain [48][49][61,62]. In contrast, the incubation period for Delta is 4 days and is shortened to 3 days for Omicron [50][63]. Both pre-symptomatic and asymptomatic transmissions have been reported [51][52][64,65]. Pre-symptomatic transmission occurs 1 to 3 days before symptom onset [53][66]. The viral load in the upper respiratory tract is highest in the first 3 to 5 days of infection [54][67], and although the loads are higher in symptomatic individuals, both pre-symptomatic and symptomatic persons can shed high viral loads, readily transmit infection and be associated with high secondary attack rates [55][68]. The viral load and duration of infectious virus shedding are higher for both the Delta and Omicron variants [56][57][69,70].
COVID-19 is a multisystem disease (Figure 3) that starts in the upper respiratory tract and lungs but can also involve the heart, blood vessels, brain, liver, kidneys and intestine [58][71]. In their majority, patients with COVID-19 present with mild symptoms consisting of fever, cough, loss of taste and/or smell and dyspnea. Other less common symptoms include headache, sore throat, chills, diarrhea, chest pain and myalgias [59][72]. Whilst most cases resolve without any serious consequences, up to 20% of cases develop moderate to severe disease needing hospitalization. This is more likely to occur in unvaccinated individuals infected with either the Alpha or Delta variants [60][61][73,74]. The case fatality rate for infected individuals is approximately 1.9% [62][75], although this may rise to 8% for people aged 70 to 79 years and to 14.8% for people 80 years and older [63][76].
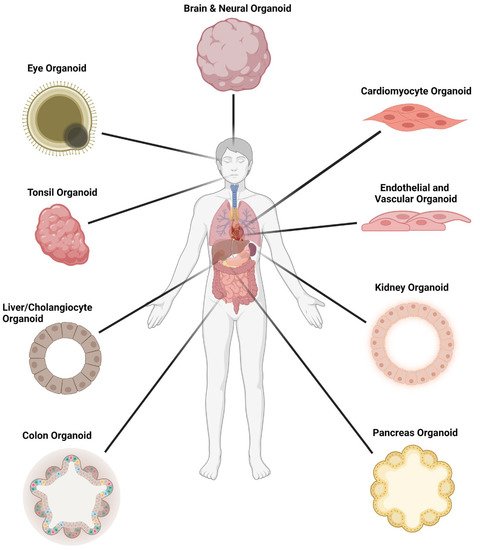

Figure 3.
Organoid models used in SARS-CoV-2 infection studies.
Figure 3
was created with
.
4. Organoid Models of SARS-CoV-2
4.1. Respiratory Tract
COVID-19 was first recognized as causing severe pneumonia; thus, it was vital to identify the regions and cell types of the respiratory tract that are infected with SARS-CoV-2. The respiratory tract architecture changes from a pseudostratified barrier epithelium lining the nose to the delicate monolayer of alveoli, which are small air sacs at the extremities of the lung enabling the exchange of oxygen and carbon dioxide (CO2) molecules into and out of the bloodstream (Figure 4). The first publications testing the respiratory cell tropism of SARS-CoV-2 used human airway epithelium (HAE) established from progenitor cells (basal cells) isolated from conductive airway tissues resected during surgery. Pseudostratified 3D tissues were differentiated at an air-liquid-interface (ALI) [64][65][84,85] or embedded in a matrix [65][85] (Figure 4). For ALI differentiation, the basal cells are seeded onto transwell membranes, and once confluent across the surface of the membrane, the apical medium is removed, exposing the cells to air. ALI-differentiation medium is refreshed in the basal chamber over 3–4 weeks to generate the pseudostratified tissue. Matrix embedded organoids are similarly seeded in an expansion phase to form cysts, and then changed to an organoid differentiation medium for 3–4 weeks [66][24][8,32]. HAE organoids are comprised of several cell types, including basal cells, ciliated cells with apical beating cilia and goblet cells secreting mucous onto the apical surface. These studies confirmed the SARS-CoV-2 infection of ciliated cells, and that viral entry and release occur from the apical surface and was soon confirmed by others [13][64][65][21,84,85]. Using a reporter virus system, RNA in situ hybridization and cultures established from different regions of the respiratory tract, Hou and colleagues [67][86] demonstrated that the ALI-differentiated nose epithelium expressed the highest levels of ACE2 and TMPRSS2 and showed the greatest SARS-CoV-2 infectivity, which gradually decreased towards the proximal respiratory tract (Figure 4). Early expression studies either did not detect or detected low levels [68][87] or did not report [65][85] ACE2 protein expression in respiratory tissues, despite robust protein expression reported in other human tissues such as the intestine analyzed in parallel [65][68][85,87]. More recent studies have confirmed ACE2 protein expression and SARS-CoV-2 infections in respiratory epithelia, identifying the differentiated ciliated cells as the dominant cell type infected [13][21][24][69][70][21,29,32,88,89] (reviewed extensively elsewhere, e.g., [71][72][73][90,91,92]).
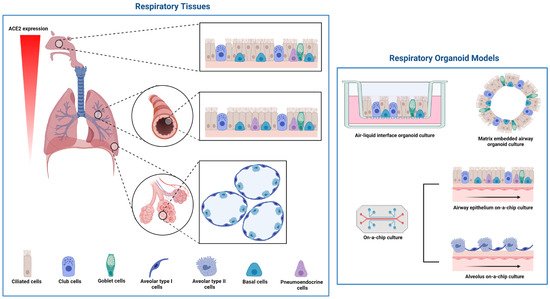

Figure 4. Cell types, and their localization within the respiratory tract, and respiratory organoid models used in SARS-CoV-2 infection studies. The morphology of respiratory tissue changes from the nose epithelium with ciliated, club, goblet, endocrine and basal cells to the delicate structure of alveoli with the flat type I cells and the secretory type II cells. Respiratory organoid culture can be tissue on a membrane (air–liquid–interface), embedded in matrix or microfluidic chip cultures. Figure 4 was created with BioRender.com.
4.2. Intestine Epithelium
The first publication using human organoids for SARS-CoV-2 infection was the result of the collaboration between the Clevers and Haagmans laboratories in the Netherlands [65][85], combining tissue-stem cell derived organoid expertise with virology. The Clevers laboratory identified Leucine-rich, repeat-containing G-protein coupled receptor 5 (Lgr5) as an exclusive maker of adult intestinal epithelial stem cells [74][100]. They showed that Lgr5+ stem cells generate the different cell types of the epithelium and self-organize into 3D structures that mimic the gut epithelium when embedded in the matrix and provided with stem cell niche growth factors [3][75][3,101]. Human small intestinal organoids (hSIOs) can be expanded indefinitely and cryopreserved, providing a renewable resource. The hSIOs can be cultured and embedded into a matrix or grown at ALI [76][9]. hSIOs [65][85] and tissues [68][87] abundantly express ACE2 and support infection by SARS-CoV and SARS-CoV-2 [65][85]. Methodologies to clone and genetically engineer hSIOs were already established in the Clevers laboratory and employed to reveal essential host factors for coronavirus infection, demonstrating the power of coupling CRISPR/Cas9 gene editing with organoids [77][33]. Moreover, the hSIOs together with human alveolar and airway ALI organoids revealed differences between the ancestral SARS-CoV-2 and the Alpha (B.1.1.7) variant. The Alpha variant produced a higher replicative fitness, producing high titers of virus later in infection [78][102].
The very high levels of ACE2 and TMPRSS2 [79][103] in hSIOs make them a permissive culture system enabling a wide infection dynamic range and a robust platform for the screening of anti-viral inhibitors. Additionally, there is potential for an oral–fecal route of transmission that requires further investigation. SARS-CoV-2 is an enveloped virus (Figure 2) and would not be expected to survive the harsh gastric environment. However, COVID-19 patients develop gastrointestinal symptoms; SARS-CoV-2 infection is reported to alter the intestinal microbiome, and SARS-CoV-2 RNA is detected in fecal samples [80][81][104,105].
4.3. Heart
Some COVID-19 patients develop severe cardiovascular complications, which led to investigations to determine if this was the direct result of the SARS-CoV-2 infection of cardiac tissue or a consequence of systemic disease. Examination by digital PCR, Western blot, immunohistochemistry, immunofluorescence, RNAscope and transmission electron microscopy assays of postmortem cardiac heart tissues of SARS-CoV-2 positive patients who showed no signs of cardiac involvement revealed that cardiomyocytes (CM) were indeed infected. Variable patterns of cardiomyocyte injury were observed [82][106]. This and other studies [83][107] examining autopsy tissues confirmed the SARS-CoV-2 presence in the heart and CM injury. To examine the human cardiac tropism of SARS-CoV-2, researchers adopted CM cultures derived from human pluripotent ES cells (hESC-CM) or human iPS cells (hiPS-CM).
The differentiation of hESCs is directed towards mesoderm lineage and then differentiation into spontaneously contracting CM, achieving 85–95% purity [84][85][108,109]. hESC-CMs express ACE2, while ES cells, mesoderm and cardiac progenitor cells do not [85][109]. hESC-CMs also express TMPRESS2, cathepsin L and furin [84][108], mirroring the expression of healthy human heart tissue (left ventricle) [84][108]. hESC-CMs are susceptible to SARS-CoV-2 infection, which is inhibited by the ACE2 antibody, camostat and E64d, indicating that cell entry requires docking to ACE2 but can proceed via an endocytic pathway (sensitive to E64d) or TMPRSS2-dependent membrane fusion (sensitive to camostat) [84][108] (Figure 2).
4.4. Liver
Several continuous cell lines were screened and utilized for SARS-CoV-2 infection experiments, including Huh7 cells, a human hepatocellular carcinoma cell line. The susceptibility of this liver cancer cell line to SARS-CoV-2 infection in vitro and the liver comorbidities associated with COVID-19 in patients with severe disease [86][113] prompted an analysis of existing single-cell (sc) RNAseq databases for cell types within the liver that express ACE2 and TMPRSS2 and could potentially be infected by SARS-CoV-2 (for example, references [87][88][114,115]). An analysis across five different liver tissue types (human fetal, healthy, cirrhotic, tumor and adjacent normal) identified a single subpopulation of human liver that co-expressed ACE2 and TMPRSS2, namely the cholangiocyte-biased progenitor pool [88][115].
Indeed, using human liver organoids, the SARS-CoV-2 infection of cholangiocytes was subsequently confirmed [89][116]. Human liver organoids established from biliary duct fragments are bipotent progenitor cells that expand as cystic organoid structures and can be differentiated towards a biliary or hepatic cell fate [90][91][117,118]. In their expansion phase, the organoid cells express cholangiocyte markers [89][91][116,118], ACE2 and TMPRSS2 and support SARS-CoV-2 infection, leading to the formation of large syncytia [89][116]. Transcriptomic analysis by scRNAseq revealed the decreased expression of genes involved in barrier functions and bile acid transport following SARS-CoV-2 infection. The advantage of human liver ductal organoids resides in their long-term expansion [91][118], providing a renewable source of primary cells for SARS-CoV-2 infection.
4.5. Brain
Initial symptoms, such as headache, dizziness and taste and smell impairment were commonly reported for SARS-CoV-2 patients, but in most cases resolved quickly. More serious indications of neurological disorder include seizures, encephalopathy and cerebrovascular disease [92][121]. MRI-based screens showed structural damage in the brains of recovering COVID-19 patients, indicating potential long-term neurological effects [93][122]. SARS-CoV-2 RNA was detected in the cerebral spinal fluid (CSF) of COVID-19 patients with neurological symptoms [94][123] and in autopsy brain samples [95][124]. To confirm or deny SARS-CoV-2 infection and identify target cell types in the brain, researchers turned to human brain organoids established from hES cells or hiPS cells. Initial results seemed conflicting, detecting SARS-CoV-2 virus in the neurons but not productive infection [96][125]. This was confirmed in a systematic test of SARS-CoV-2 neurotropism in region-specific brain organoids (cortical, hippocampal, hypothalamic and mid-brain), which also did not show productive SARS-CoV-2 infection [97][126]. Subsequent studies with brain organoids that contained choroid plexus epithelium were productively infected, but only in the choroid plexus region of the organoid [97][98][126,127]. Tropism for choroid plexus epithelium was confirmed using pure choroid plexus organoids [97][126]. It is proposed that productive SARS-CoV-2 infection leads to the damage of the epithelium and loss of barrier function, allowing viral entry into the central nervous system (CNS).
Another potential portal of entry into the CNS is from the nasal cavity through the olfactory bulb. An elegant study by Meinhardt and colleagues [99][128] showed that SARS-CoV-2 can enter the CNS by crossing the neural-mucosal interface in olfactory mucosa. The proximity of mucosal, endothelial and nervous tissue, including olfactory and sensory nerve endings, coupled with the robust SARS-CoV-2 infection of the nasal epithelium, might precipitate entry into the CNS via the olfactory tract. This may explain some of the symptoms of COVID-19, including loss of smell and taste.
4.6. Kidney
ACE2 is strongly expressed in kidney tubules [68][100][101][87,129,130]; SARS-CoV-2 can be detected in urine [102][131] and in autopsy kidney tissue from SARS-CoV-2-infected patients [103][104][132,133], and COVID-19 patients show signs of kidney damage [58][105][106][71,134,135]. To determine if the kidney function decline reported for COVID-19 patients could be due to the infection of kidney cells by SARS-CoV-2, hES-derived kidney epithelium organoids were interrogated. The engineered kidney organoids expressed ACE2 and TMPRSS2 and were infected with SARS-CoV-2, producing infectious virus, and the infection was inhibited by soluble ACE2 [107][108][136,137]. Remdesivir, an RNA polymerase inhibitor that blocks viral replication, had an additive anti-viral effect when combined with soluble ACE2 in the kidney organoids [109][138].
4.7. The Eye—An Alternative Route of Entry
Although the respiratory system is considered the primary route for SARS-CoV-2 infection, and the nose epithelium the dominant initial site of infection, observations by ophthalmologists early in the pandemic raised the possibility that the eye was also a route of viral entry [110][141]. People contracted COVID-19 despite wearing a face mask to protect the respiratory route of entry, which led to investigations of the ocular surface as a site of infection by SARS-CoV-2. The ocular surface includes the cornea, sclera and limbus, and an examination of post-mortem samples demonstrated that these tissues express ACE2 and TMPRSS2 [111][112][142,143] and were positive for SARS-CoV-2 S protein by immunofluorescence in samples from SARS-CoV-2 positive cadavers [111][142]. To confirm that these tissue cell types were indeed susceptible to SARS-CoV-2 infection, cultures were established from cells isolated from eye regions of healthy donors and infected with SARS-CoV-2. The limbus cells were the most susceptible primary ocular tissue to SARS-CoV-2 infection [111][142]. Accessing human eye tissue relies on organ donation, and the cultures established have a limited lifespan and complexity. Human ES cell and iPS cell-derived retinal [113][144] and “whole eye” [111][114][142,145] organoids have been generated and confirm endogenous ACE2 and TMPRSS2 expression [111][115][142,146], and that SARS-CoV-2 infection is primarily in limbus-like cells [111][114][142,145]. Collectively, these data demonstrate the power of pluripotent stem cell-derived organoid technology in establishing the eye as a potential mode of SARS-CoV-2 transmission and emphasizes the importance of face shields to prevent touching the face or eye, especially in the case of conjunctivitis.
4.8. Vasculature
The vasculature, like neuronal networks, is of great importance to the understanding of COVID-19 pathogenesis as it encompasses all organs. The endothelium lines the inner surface of all blood vessels and plays crucial roles in human physiology and pathophysiology, including COVID-19. The endothelium functions in both adaptive and innate immunity; it provides an anticoagulant surface and maintains blood vessel barrier integrity. The dysregulation of all these functions features in the sequelae of COVID-19 (reviewed extensively, e.g., [116][147]). However, it remains controversial whether endothelial damage is a direct effect of SARS-CoV-2 infection of the endothelial cells.
Given that severe COVID-19 is a multi-organ disease, the initial focus was on the affected organs. Nonetheless, endothelial damage is causally implicated in COVID-19 organ dysfunction (reviewed extensively, e.g., [116][117][118][119][120][147,148,149,150,151]). Early in the pandemic, it was recognized that circulating endothelial cells in COVID-19 patients, which is indicative of endothelial damage, correlated with increases in markers of disease severity and inflammatory cytokines [121][122][152,153]. Examinations of autopsy tissues from COVID-19 patients by immunohistochemistry revealed an accumulation of inflammatory cells associated with the endothelium in vascular beds of several organs, including the heart, small intestine and lung [123][154]. Further examination of kidney post-mortem tissues by electron microscopy showed viral inclusion bodies in the peritubular space and viral particles in endothelial cells of the glomerular capillary loops [123][154]. The productive SARS-CoV-2 infection of endothelial cells was confirmed using hiPS-derived capillary organoids. Infection of these blood vessel organoids was inhibited by soluble human ACE2, indicating ACE2-mediated viral entry [107][136]. In a similar vein, Yang and colleagues [124][155] used hES to generate eight different distinct cell types or organoids modelling the embryonic development of tissues derived from the three germ layers (definitive endoderm, mesoderm and ectoderm). Productive SARS-CoV-2 infection was confirmed in several tissue types, including endothelial cells [124][155]. Mouse models of SARS-CoV-2 also support the direct infection of lung and cardiac endothelial cells [125][156].
Nonetheless, the extent to which the direct infection of endothelial cells by SARS-CoV-2 contributes to vascular involvement in COVID-19 remains to be determined and may indeed be organ and tissue dependent. In vitro cultures of primary human umbilical vein endothelial cells (HUVECs) and human microvascular endothelial cells from the lung (HMVEC-L) did not support SARS-CoV-2 infection [126][157].
4.9. Other Organs
Many other organoids derived from tissue stem cells or human pluripotent stem cells have been adopted for SARS-CoV-2 research to define cell tropism and to understand pathogenesis and modes of transmission (Figure 3). These include tonsils and the oral cavity, with saliva as a potential means of transmission, the pancreas, alveolar macrophages and gastric epithelium, to name a few (reviewed comprehensively elsewhere in addition to review articles cited here).
