Oesophageal cancer (OC) is the ninth most common cancer worldwide. Patients receive neoadjuvant therapy (NAT) as standard of care, but less than 20% of patients with oesophageal adenocarcinoma (OAC) or a third of oesophageal squamous cell carcinoma (OSCC) patients, obtain a clinically meaningful response. Developing a method of determining a patient’s response to NAT before treatment will allow rational treatment decisions to be made, thus improving patient outcome and quality of life. MicroRNAs are valuable biomarkers of response to NAT in OC. Research is needed to understand the effects different types of chemotherapy and chemoradiotherapy have on the predictive value of microRNAs; studies also require greater standardization in how response is defined.
- oesophageal adenocarcinoma
- oesophageal squamous cell carcinoma
- predicting response
- chemotherapy
- chemoradiotherapy
- neoadjuvant therapy
- microRNAs
1. Introduction
1.1. Oesophageal Cancer Epidemiology
1.2. Pathophysiology of OAC and OSCC
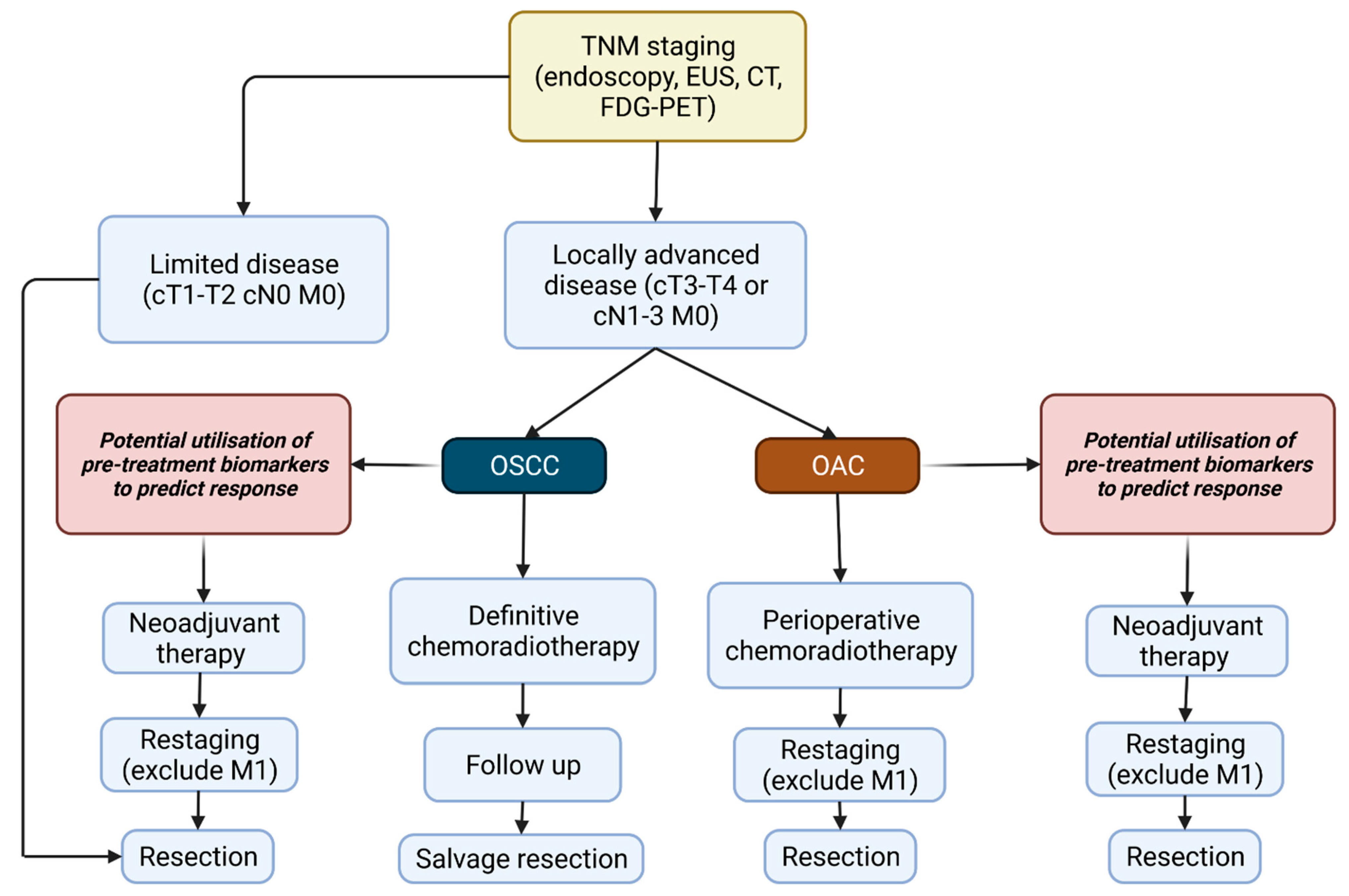
1.3. Current Pathways of Screening
1.4. Treatment of Oesophageal Cancer
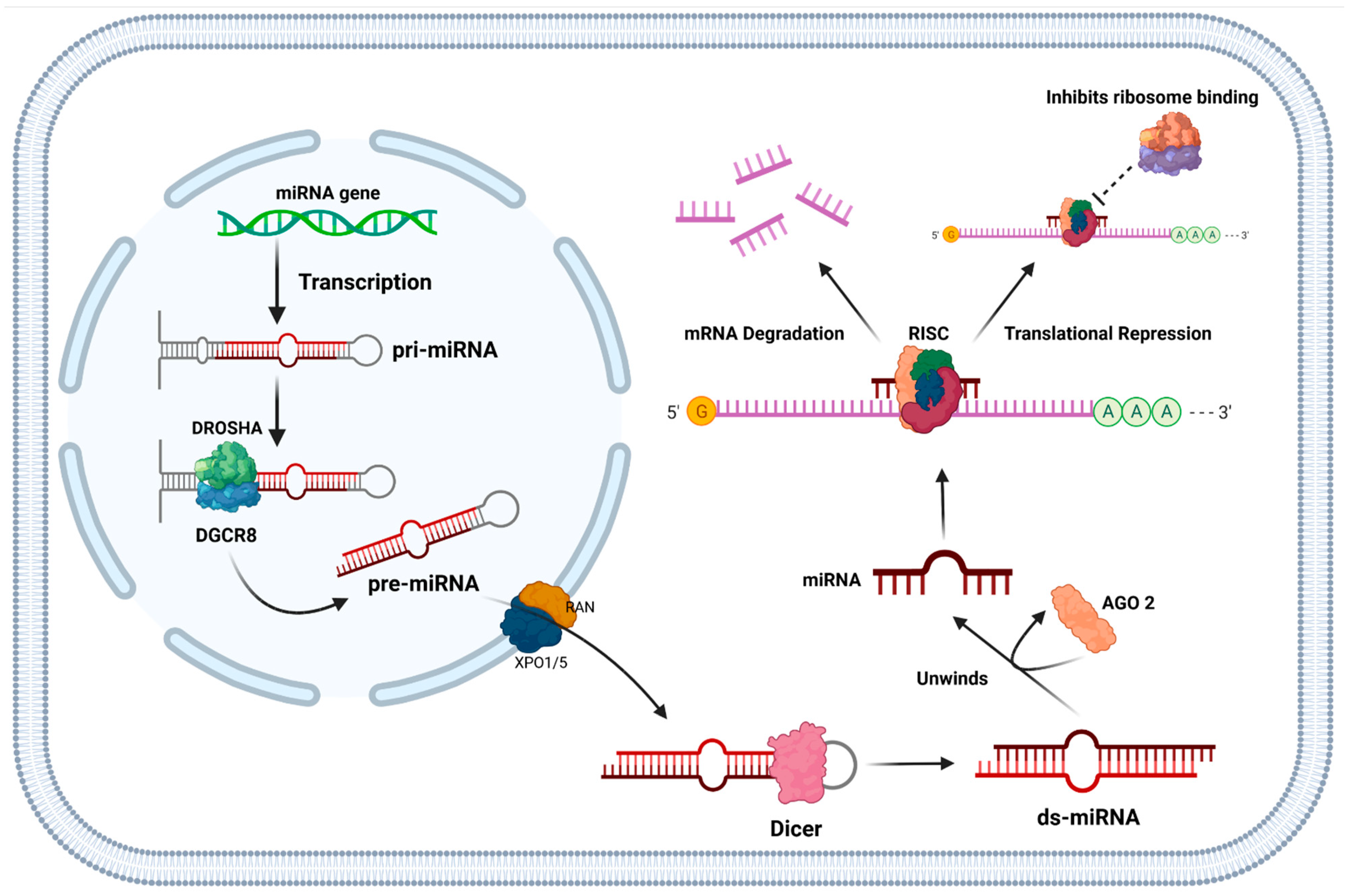
1.5. Function of miRNAs and Their Role in Cancer
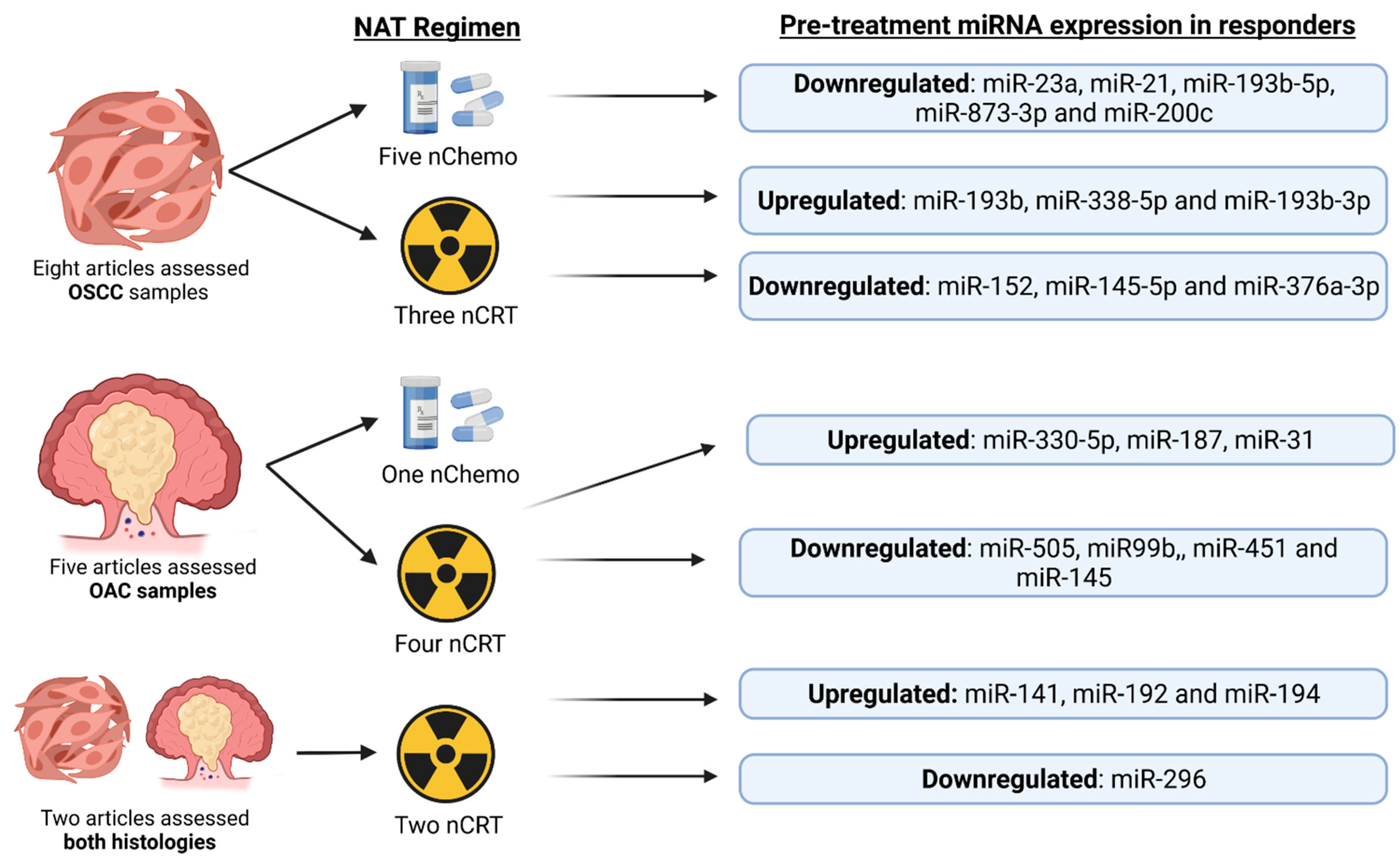
2. Current Insights
Current Use of miR-21, miR-193b and miR-200c in Cancers and Their Functional Roles
References
- Esophageal Cancer: Practice Essentials, Background, Anatomy. 2020. Available online: https://emedicine.medscape.com/article/277930-overview (accessed on 9 November 2020).
- Kamangar, F.; Nasrollahzadeh, D.; Safiri, S.; Sepanlou, S.G.; Fitzmaurice, C.; Ikuta, K.S.; Bisignano, C.; Islami, F.; Roshandel, G.; Lim, S.S.; et al. The Global, Regional, and National Burden of Oesophageal Cancer and Its Attributable Risk Factors in 195 Countries and Territories, 1990–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 582–597.
- Roser, M.; Ritchie, H. Cancer. Our World Data. 2015. Available online: https://ourworldindata.org/cancer (accessed on 9 November 2020).
- Oesophageal Cancer Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/oesophageal-cancer (accessed on 9 November 2020).
- Cancer Survival Rates. Available online: https://www.nuffieldtrust.org.uk/resource/cancer-survival-rates (accessed on 9 November 2020).
- Early Diagnosis. Available online: https://crukcancerintelligence.shinyapps.io/EarlyDiagnosis/ (accessed on 16 January 2022).
- Smyth, E.C.; Lagergren, J.; Fitzgerald, R.C.; Lordick, F.; Shah, M.A.; Lagergren, P.; Cunningham, D. Oesophageal Cancer. Nat. Rev. Dis. Primers 2017, 3, 17048.
- Runge, T.M.; Abrams, J.A.; Shaheen, N.J. Epidemiology of Barrett’s Esophagus and Esophageal Adenocarcinoma. Gastroenterol. Clin. N. Am. 2015, 44, 203–231.
- Wong, I.; Law, S. The CROSS Road in Neoadjuvant Therapy for Esophageal Cancer: Long-Term Results of CROSS Trial. Transl. Cancer Res. 2016, 3, S415–S419.
- Ritchie, H.; Roser, M. Smoking. Our World Data. 2013. Available online: https://ourworldindata.org/smoking#:~:text=Smoking%20is%20one%20the%20leading,1%2Din%2D5%20deaths.&text=Nearly%20one%2Din%2Dfour%20adults%20in%20the%20world%20smoke%20tobacco (accessed on 9 November 2020).
- Ramachandran, A.; Chamukuttan, S.; Shetty, S.A.; Arun, N.; Susairaj, P. Obesity in Asia—Is It Different from Rest of the World. Diabetes Metab. Res. Rev. 2012, 28, 47–51.
- Oesophageal Cancers Country Data. Available online: https://digestivecancers.eu/oesophageal-cancer-map/ (accessed on 9 November 2020).
- di Pietro, M.; Alzoubaidi, D.; Fitzgerald, R.C. Barrett’s Esophagus and Cancer Risk: How Research Advances Can Impact Clinical Practice. Gut Liver 2014, 8, 356–370.
- Fléjou, J.-F. Barrett’s Oesophagus: From Metaplasia to Dysplasia and Cancer. Gut 2005, 54, i6–i12.
- Weaver, J.M.J.; Ross-Innes, C.S.; Shannon, N.; Lynch, A.G.; Forshew, T.; Barbera, M.; Murtaza, M.; Ong, C.-A.J.; Lao-Sirieix, P.; Dunning, M.J.; et al. Ordering of Mutations in Preinvasive Disease Stages of Esophageal Carcinogenesis. Nat. Genet. 2014, 46, 837–843.
- Esophageal Cancer—Symptoms and Causes. Available online: https://www.mayoclinic.org/diseases-conditions/esophageal-cancer/symptoms-causes/syc-20356084 (accessed on 3 December 2020).
- Stewart, B.W.; Wild, C.P. (Eds.) World Cancer Report 2014; World Health Organization: Geneva, Switzerland, 2014; ISBN 978-92-832-0432-9.
- Testa, U.; Castelli, G.; Pelosi, E. Esophageal Cancer: Genomic and Molecular Characterization, Stem Cell Compartment and Clonal Evolution. Medicines 2017, 4, 67.
- Screening | Oesophageal Cancer | Cancer Research, UK. Available online: https://www.cancerresearchuk.org/about-cancer/oesophageal-cancer/getting-diagnosed/screening (accessed on 16 December 2020).
- Bollschweiler, E.; Boettcher, K.; Hoelscher, A.H.; Sasako, M.; Kinoshita, T.; Maruyama, K.; Siewert, J.R. Is the Prognosis for Japanese and German Patients with Gastric Cancer Really Different? Cancer 1993, 71, 2918–2925.
- Zakko, L.; Lutzke, L.; Wang, K.K. Screening and Preventive Strategies in Esophagogastric Cancer. Surg. Oncol. Clin. 2017, 26, 163–178.
- Ben-Menachem, T.; Decker, G.A.; Early, D.S.; Evans, J.; Fanelli, R.D.; Fisher, D.A.; Fisher, L.; Fukami, N.; Hwang, J.H.; Ikenberry, S.O.; et al. Adverse Events of Upper GI Endoscopy. Gastrointest. Endosc. 2012, 76, 707–718.
- Chen, R.; Liu, Y.; Song, G.; Li, B.; Zhao, D.; Hua, Z.; Wang, X.; Li, J.; Hao, C.; Zhang, L.; et al. Effectiveness of One-Time Endoscopic Screening Programme in Prevention of Upper Gastrointestinal Cancer in China: A Multicentre Population-Based Cohort Study. Gut 2020, 70, 251–260.
- Ross-Innes, C.S.; Debiram-Beecham, I.; O’Donovan, M.; Walker, E.; Varghese, S.; Lao-Sirieix, P.; Lovat, L.; Griffin, M.; Ragunath, K.; Haidry, R.; et al. Evaluation of a Minimally Invasive Cell Sampling Device Coupled with Assessment of Trefoil Factor 3 Expression for Diagnosing Barrett’s Esophagus: A Multi-Center Case-Control Study. PLoS Med. 2015, 12, e1001780.
- Fitzgerald, R.C.; Di Pietro, M.; O’Donovan, M.; Maroni, R.; Muldrew, B.; Debiram-Beecham, I.; Gehrung, M.; Offman, J.; Tripathi, M.; Smith, S.G.; et al. Cytosponge-Trefoil Factor 3 versus Usual Care to Identify Barrett’s Oesophagus in a Primary Care Setting: A Multicentre, Pragmatic, Randomised Controlled Trial. Lancet 2020, 396, 333–344.
- Oesophageal and Gastric Cancer Overview—NICE Pathways. Available online: https://pathways.nice.org.uk/pathways/oesophageal-and-gastric-cancer#content=view-node%3Anodes-staging (accessed on 3 December 2020).
- Lordick, F.; Mariette, C.; Haustermans, K.; Obermannová, R.; Arnold, D. Oesophageal Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2016, 27, v50–v57.
- Cunningham, D.; Allum, W.H.; Stenning, S.P.; Thompson, J.N.; Van de Velde, C.J.H.; Nicolson, M.; Scarffe, J.H.; Lofts, F.J.; Falk, S.J.; Iveson, T.J.; et al. Perioperative Chemotherapy versus Surgery Alone for Resectable Gastroesophageal Cancer. N. Engl. J. Med. 2006, 355, 11–20.
- van Hagen, P.; Hulshof, M.C.C.M.; van Lanschot, J.J.B.; Steyerberg, E.W.; Henegouwen, M.V.B.; Wijnhoven, B.P.L.; Richel, D.J.; Nieuwenhuijzen, G.A.P.; Hospers, G.A.P.; Bonenkamp, J.J.; et al. Preoperative Chemoradiotherapy for Esophageal or Junctional Cancer. N. Engl. J. Med. 2012, 366, 2074–2084.
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. MicroRNAs as Oncogenes and Tumor Suppressors. Dev. Biol. 2007, 302, 1–12.
- Noble, F.; Lloyd, M.A.; Turkington, R.; Griffiths, E.; O’Donovan, M.; O’Neill, J.R.; Mercer, S.; Parsons, S.L.; Fitzgerald, R.C.; Underwood, T.J.; et al. Multicentre Cohort Study to Define and Validate Pathological Assessment of Response to Neoadjuvant Therapy in Oesophagogastric Adenocarcinoma. Br. J. Surg. 2017, 104, 1816–1828.
- Lordick, F.; Gockel, I. Chances, Risks and Limitations of Neoadjuvant Therapy in Surgical Oncology. Innov. Surg. Sci. 2016, 1, 3–11.
- Barbour, A.P.; Walpole, E.T.; Mai, G.T.; Barnes, E.H.; Watson, D.I.; Ackland, S.P.; Martin, J.M.; Burge, M.; Finch, R.; Karapetis, C.S.; et al. Preoperative Cisplatin, Fluorouracil, and Docetaxel with or without Radiotherapy after Poor Early Response to Cisplatin and Fluorouracil for Resectable Oesophageal Adenocarcinoma (AGITG DOCTOR): Results from a Multicentre, Randomised Controlled Phase II Trial. Ann. Oncol. 2020, 31, 236–245.
- Pathologic Assessment of Tumor Regression after Preoperative Chemoradiotherapy of Esophageal Carcinoma. Clinicopathologic Correlations—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/8194005/ (accessed on 26 January 2022).
- Wang, D.; Smit, J.K.; Zwaan, E.; Muijs, C.T.; Groen, H.; Hollema, H.; Plukker, J.T. Neoadjuvant Therapy Reduces the Incidence of Nodal Micrometastases in Esophageal Adenocarcinoma. Am. J. Surg. 2013, 206, 732–738.
- Hiraki, Y.; Kimura, Y.; Imano, M.; Kato, H.; Iwama, M.; Shiraishi, O.; Yasuda, A.; Shinkai, M.; Makino, T.; Motoori, M.; et al. Controlling Lymph Node Micrometastases by Neoadjuvant Chemotherapy Affects the Prognosis in Advanced Esophageal Squamous Cell Carcinoma. Surg. Today 2021, 51, 118–126.
- Ameres, S.L.; Zamore, P.D. Diversifying MicroRNA Sequence and Function. Nat. Rev. Mol. Cell Biol. 2013, 14, 475–488.
- Lai, E.C. Micro RNAs Are Complementary to 3′ UTR Sequence Motifs That Mediate Negative Post-Transcriptional Regulation. Nat. Genet. 2002, 30, 363–364.
- Zhang, Y. RNA-Induced Silencing Complex (RISC). In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; p. 1876. ISBN 978-1-4419-9863-7.
- Ding, L.; Lan, Z.; Xiong, X.; Ao, H.; Feng, Y.; Gu, H.; Yu, M.; Cui, Q. The Dual Role of MicroRNAs in Colorectal Cancer Progression. Int. J. Mol. Sci. 2018, 19, 2791.
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561.
- Essakly, A.; Loeser, H.; Kraemer, M.; Alakus, H.; Chon, S.-H.; Zander, T.; Buettner, R.; Hillmer, A.M.; Bruns, C.J.; Schroeder, W.; et al. PIK3CA and KRAS Amplification in Esophageal Adenocarcinoma and Their Impact on the Inflammatory Tumor Microenvironment and Prognosis. Transl. Oncol. 2019, 13, 157–164.
- Xu, J.; Lv, H.; Zhang, B.; Xu, F.; Zhu, H.; Chen, B.; Zhu, C.; Shen, J. MiR-30b-5p Acts as a Tumor Suppressor MicroRNA in Esophageal Squamous Cell Carcinoma. J. Thorac. Dis. 2019, 11, 3015–3029.
- Cui, X.-B.; Peng, H.; Li, R.-R.; Mu, J.-Q.; Yang, L.; Li, N.; Liu, C.-X.; Hu, J.-M.; Li, S.-G.; Wei, Y.; et al. MicroRNA-34a Functions as a Tumor Suppressor by Directly Targeting Oncogenic PLCE1 in Kazakh Esophageal Squamous Cell Carcinoma. Oncotarget 2017, 8, 92454–92469.
- Skinner, H.D.; Lee, J.H.; Bhutani, M.S.; Weston, B.; Hofstetter, W.; Komaki, R.; Shiozaki, H.; Wadhwa, R.; Sudo, K.; Elimova, E.; et al. A Validated MiRNA Profile Predicts Response to Therapy in Esophageal Adenocarcinoma. Cancer 2014, 120, 3635–3641.
- Lindholm, E.M.; Aure, M.R.; Haugen, M.H.; Sahlberg, K.K.; Kristensen, V.N.; Nebdal, D.; Børresen-Dale, A.-L.; Lingjærde, O.C.; Engebraaten, O. MiRNA Expression Changes during the Course of Neoadjuvant Bevacizumab and Chemotherapy Treatment in Breast Cancer. Mol. Oncol. 2019, 13, 2278–2296.
- Kheirelseid, E.A.H.; Miller, N.; Chang, K.H.; Curran, C.; Hennessey, E.; Sheehan, M.; Newell, J.; Lemetre, C.; Balls, G.; Kerin, M.J. MiRNA Expressions in Rectal Cancer as Predictors of Response to Neoadjuvant Chemoradiation Therapy. Int. J. Colorectal Dis. 2013, 28, 247–260.
- Gao, S.; Zhao, Z.-Y.; Zhang, Z.-Y.; Zhang, Y.; Wu, R. Prognostic Value of MicroRNAs in Esophageal Carcinoma: A Meta-Analysis. Clin. Transl. Gastroenterol. 2018, 9, 203.
- Wen, J.; Luo, K.; Liu, H.; Liu, S.; Lin, G.; Hu, Y.; Zhang, X.; Wang, G.; Chen, Y.; Chen, Z.; et al. MiRNA Expression Analysis of Pretreatment Biopsies Predicts the Pathological Response of Esophageal Squamous Cell Carcinomas to Neoadjuvant Chemoradiotherapy. Ann. Surg. 2016, 263, 942–948.
- Rahman, S.A.; Walker, R.C.; Lloyd, M.A.; Grace, B.L.; van Boxel, G.I.; Kingma, B.F.; Ruurda, J.P.; van Hillegersberg, R.; Harris, S.; Parsons, S.; et al. Machine Learning to Predict Early Recurrence after Oesophageal Cancer Surgery. Br. J. Surg. 2020, 107, 1042–1052.
- Pilonis, N.D.; Killcoyne, S.; Tan, W.K.; O’Donovan, M.; Malhotra, S.; Tripathi, M.; Miremadi, A.; Debiram-Beecham, I.; Evans, T.; Phillips, R.; et al. Use of a Cytosponge Biomarker Panel to Prioritise Endoscopic Barrett’s Oesophagus Surveillance: A Cross-Sectional Study Followed by a Real-World Prospective Pilot. Lancet Oncol. 2022, 23, 270–278.
- Drucker, E.; Krapfenbauer, K. Pitfalls and Limitations in Translation from Biomarker Discovery to Clinical Utility in Predictive and Personalised Medicine. EPMA J. 2013, 4, 7.
- Brown, R.A.M.; Epis, M.R.; Horsham, J.L.; Kabir, T.D.; Richardson, K.L.; Leedman, P.J. Total RNA Extraction from Tissues for MicroRNA and Target Gene Expression Analysis: Not All Kits Are Created Equal. BMC Biotechnol. 2018, 18, 16.
- Moldovan, L.; Batte, K.E.; Trgovcich, J.; Wisler, J.; Marsh, C.B.; Piper, M. Methodological Challenges in Utilizing MiRNAs as Circulating Biomarkers. J. Cell. Mol. Med. 2014, 18, 371–390.
- Atarod, S.; Smith, H.; Dickinson, A.; Wang, X.-N. MicroRNA Levels Quantified in Whole Blood Varies from PBMCs. F1000Research 2014, 3, 183.
- Bibby, B.A.S.; Reynolds, J.V.; Maher, S.G. MicroRNA-330-5p as a Putative Modulator of Neoadjuvant Chemoradiotherapy Sensitivity in Oesophageal Adenocarcinoma. PLoS ONE 2015, 10, e0134180.
- Jiang, W.; de Jong, J.M.; van Hillegersberg, R.; Read, M. Predicting Response to Neoadjuvant Therapy in Oesophageal Adenocarcinoma. Cancers 2022, 14, 996.
- Li, F.; Lv, J.-H.; Liang, L.; Wang, J.; Li, C.-R.; Sun, L.; Li, T. Downregulation of MicroRNA-21 Inhibited Radiation-Resistance of Esophageal Squamous Cell Carcinoma. Cancer Cell Int. 2018, 18, 39.
- Nouraee, N.; Van Roosbroeck, K.; Vasei, M.; Semnani, S.; Samaei, N.M.; Naghshvar, F.; Omidi, A.A.; Calin, G.A.; Mowla, S.J. Expression, Tissue Distribution and Function of MiR-21 in Esophageal Squamous Cell Carcinoma. PLoS ONE 2013, 8, e73009.
- Kestens, C.; Siersema, P.D.; van Baal, J.W. Current Understanding of the Functional Roles of Aberrantly Expressed MicroRNAs in Esophageal Cancer. World J. Gastroenterol. 2016, 22, 1–7.
- Ourô, S.; Mourato, C.; Velho, S.; Cardador, A.; Ferreira, M.P.; Albergaria, D.; Castro, R.E.; Maio, R.; Rodrigues, C.M.P. Potential of MiR-21 to Predict Incomplete Response to Chemoradiotherapy in Rectal Adenocarcinoma. Front. Oncol. 2020, 10, 2212.
- Liu, B.; Su, F.; Lv, X.; Zhang, W.; Shang, X.; Zhang, Y.; Zhang, J. Serum MicroRNA-21 Predicted Treatment Outcome and Survival in HER2-Positive Breast Cancer Patients Receiving Neoadjuvant Chemotherapy Combined with Trastuzumab. Cancer Chemother. Pharmacol. 2019, 84, 1039–1049.
- Fang, Z.; Li, C.; Li, S. MicroRNA-193b Acts as a Tumor Suppressor in Colon Cancer Progression via Targeting RAB22A. Exp. Ther. Med. 2019, 17, 3921–3928.
- Nyhan, M.J.; O’Donovan, T.R.; Boersma, A.W.M.; Wiemer, E.A.C.; McKenna, S.L. MiR-193b Promotes Autophagy and Non-Apoptotic Cell Death in Oesophageal Cancer Cells. BMC Cancer 2016, 16, 101.
- Hummel, R.; Sie, C.; Watson, D.I.; Wang, T.; Ansar, A.; Michael, M.Z.; Van der Hoek, M.; Haier, J.; Hussey, D.J. MicroRNA Signatures in Chemotherapy Resistant Esophageal Cancer Cell Lines. World J. Gastroenterol. WJG 2014, 20, 14904–14912.
- Czochor, J.R.; Glazer, P.M. MicroRNAs in Cancer Cell Response to Ionizing Radiation. Antioxid. Redox Signal. 2014, 21, 293–312.
- Chan, C.; Lai, K.; Ng, E.; Kiang, M.; Kwok, T.; Wang, H.; Chan, K.; Law, T.; Tong, D.; Chan, K.; et al. Serum MicroRNA-193b as a Promising Biomarker for Prediction of Chemoradiation Sensitivity in Esophageal Squamous Cell Carcinoma Patients. Oncol. Lett. 2017, 15, 3273–3280.
- Tanaka, K.; Miyata, H.; Yamasaki, M.; Sugimura, K.; Takahashi, T.; Kurokawa, Y.; Nakajima, K.; Takiguchi, S.; Mori, M.; Doki, Y. Circulating MiR-200c Levels Significantly Predict Response to Chemotherapy and Prognosis of Patients Undergoing Neoadjuvant Chemotherapy for Esophageal Cancer. Ann. Surg. Oncol. 2013, 20, 607–615.
- Mahawongkajit, P.; Tomtitchong, P. Expression of MiRNA in 5-FU Resistant Esophageal Cancer. Mol. Clin. Oncol. 2020, 13, 221–227.
- Sulaiman, S.A.; Ab Mutalib, N.-S.; Jamal, R. MiR-200c Regulation of Metastases in Ovarian Cancer: Potential Role in Epithelial and Mesenchymal Transition. Front. Pharmacol. 2016, 7, 271.
- Smith, C.M.; Watson, D.I.; Leong, M.P.; Mayne, G.C.; Michael, M.Z.; Wijnhoven, B.P.; Hussey, D.J. MiR-200 Family Expression Is Downregulated upon Neoplastic Progression of Barrett’s Esophagus. World J. Gastroenterol. WJG 2011, 17, 1036–1044.
- Hamano, R.; Miyata, H.; Yamasaki, M.; Kurokawa, Y.; Hara, J.; Moon, J.H.; Nakajima, K.; Takiguchi, S.; Fujiwara, Y.; Mori, M.; et al. Overexpression of MiR-200c Induces Chemoresistance in Esophageal Cancers Mediated Through Activation of the Akt Signaling Pathway. Clin. Cancer Res. 2011, 17, 3029–3038.
