Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Lindsay Dong and Version 2 by Lindsay Dong.
Malaria is a vector-transmitted parasite disease that continues to plague mankind. It is caused in humans by five main species of Plasmodium. The majority of conventional antimalarials kill parasites via direct or indirect overproduction of reactive oxygen species (ROS). Even when some parasites try to manage these ROS, over production of the ROS still leads to parasite death. This therefore underscores the role of ROS in the antiplasmodial activity of different antimalarials.
- ROS
- antimalarials
- Plasmodium falciparum
- malaria and oxidative stress
1. ROS in Living Cells
ROS are defined as oxygen-containing reactive species. This term includes superoxide radicals (O
2•−
), hydrogen peroxide (H
2
O
2
), hydroxyl radicals (
•
OH), singlet oxygen (
1
O
2
), peroxyl radicals (LOO
•
), alkoxyl radicals (LO
•
), lipid hydroperoxide (LOOH), and peroxynitrite (ONOO
−
), among others
[1]
. Their generation can first occur as a result of the partial reduction of oxygen, as described in
Figure 1
. Among the ROS, some species are radicals, i.e., have unpaired electrons in their outer orbit—for example, superoxide and hydroxyl radicals. The different forms of ROS have varying levels of reactivity depending on their oxidation potential, with H
2
O
2
being the least reactive and
▪OH being the most reactive.
OH being the most reactive.
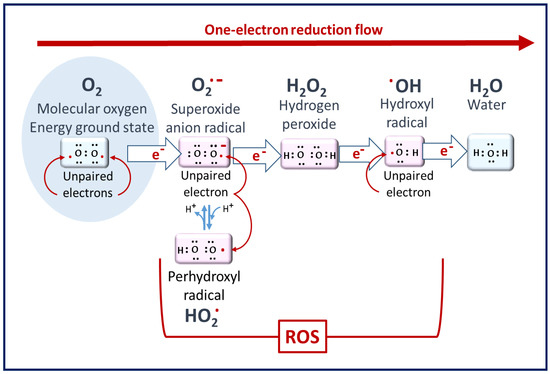
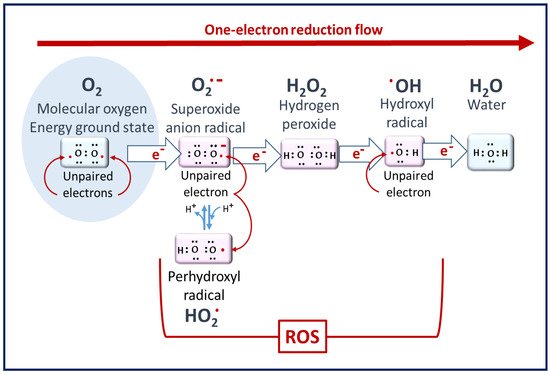
Figure 1. Molecular oxygen to ROS. Superoxide radicals are the primary ROS but have poor reactivity. Hydrogen peroxide is the least reactive ROS. At the end of the reduction flow, hydroxyl radicals are the most reactive [2].
2. The Biochemical Impacts of ROS in Living Cells
ROS generated at low concentrations under physiological conditions are often beneficial, playing important roles as regulatory mediators in signaling processes [1][3]. Nevertheless, at high concentrations, they become deleterious for the cells. The toxicity of free radicals stems from their unstable nature and predisposition to donate or abstract electrons from nearby molecules, triggering a chain reaction that can be terminated by another molecule with unpaired electrons. Lipids, proteins, and nucleic acids are attacked and damaged by ROS, which affects the survival of living organisms [2][1] (Figure 2).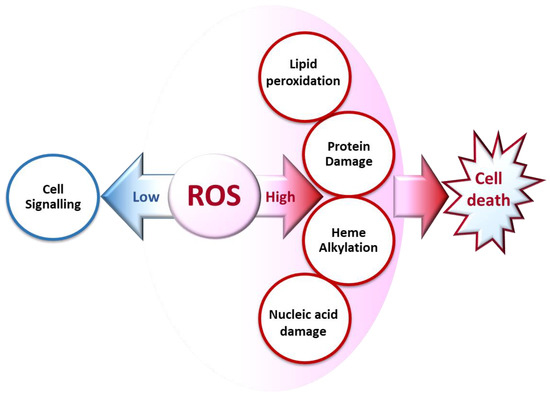
Figure 2. Biochemical impacts of ROS depending on their concentrations.
2.1. Cell Signaling
ROS play important physiological roles, such as in the induction of apoptosis and suppression of some genes’ expression, at low concentrations [3]. Studies have reported the production of ROS by specialized plasma membrane oxidases and nicotinamide adenine dinucleotide phosphate (NADPH) oxidases in normal physiological signaling by growth factors and cytokines [8].
2.2. Lipid Peroxidation
Lipids are prone to attacks from reactive species, resulting in their oxidation and the formation of lipid peroxides [9] (Figure 3). This leads to cellular dysfunction, especially as lipids are major components of cell membranes. In Plasmodium, artemisinin and its derivatives accumulate in neutral lipid bodies, especially in the digestive vacuole, where they can trigger oxidative damage after their heme–iron activation [10], via a lipid peroxidation process that, once initiated, is propagated by autocatalysis to free fatty acids [10]. This oxidative damage leads to a loss of Plasmodium membrane integrity and, consequently, parasite death. Moreover, tetraoxanes oxidatively damage phospholipids more than artemisinin [11], which may account for the differential antimalarial effect of these endoperoxides.
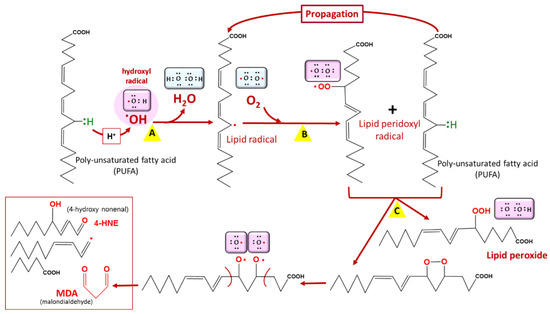
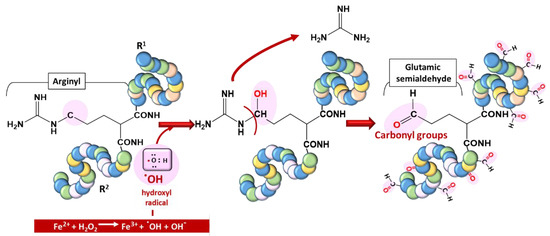

Figure 3. Lipid peroxidation. Lipid peroxidation occurs in three phases: initiation (A), propagation (B), and termination (C). Malondialdehyde (MDA) is a biomarker of lipid peroxidation in living cells. Among lipids, polyunsaturated fatty acids (PUFAs) are the most vulnerable to lipid peroxidation. COOH = carboxyl group, OOH = hydroperoxyl.
2.3. Protein Damage
Proteins are vulnerable to oxidation by ROS, a phenomenon generally referred to as protein carbonylation. Protein carbonyls are reactive aldehydes and ketone adducts formed through the α-amidation pathway, the formation of protein–protein cross-linked derivatives, the oxidative cleavage of glutamyl residues, and cell membrane damage by lipid oxidation products [12]. Protein carbonyls have been used as biomarkers of oxidative stress due to their relative stability and early formation [13]. ROS generate cytotoxic protein carbonyls in two main and irreversible ways: (i) by a metal-catalyzed oxidative (MCO) attack targeting the amino acid moiety of arginine, lysine, threonine, and proline (Figure 4);
Figure 4. Typical carbonylation of proteins based on a metal-catalyzed oxidation (MCO) attack involving a transition metal (Fe2+) for hydroxyl radical generation.
(ii) by secondary reactions on lysine, cysteine, and histidine with reactive carbonyl derivatives, resulting, among other things, from lipid peroxidation.
These post-translational modifications due to the oxidization of amino acid side chains, which results in aldehyde, ketone, and lactam formation, can also lead to damage to proteins [14][15].The endoperoxides alkylate numerous vital proteins, including enzymes in the parasite, particularly the cysteine residue of cysteine proteases, which play a vast role in P. falciparum, ranging from hemoglobin (Hb) uptake and digestion to aiding red blood cell rupture [16][17][18]. Therefore, their alteration, leading to the inhibition of their enzymatic properties, can cause severe setbacks for parasite survival, considering the huge dependence of the parasite on Hb digestion products. Endoperoxides also disrupt the activities of sarcoplasmic–endoplasmic reticulum Ca2+-ATPase- (SERCA-) type protein, encoded by the pfatpase6 gene, presumably from the ROS that they generate [19]. More recently, approximately 124 proteins were identified as covalent binding targets of artemisinins [20].
2.4. Nucleic Acid Damage
The molecular integrity of DNA and other genetic material is necessary for the continued existence and survival of all living organisms, including Plasmodium. ROS cause structural damage to DNA by attacking mainly one of its bases, guanine, due to its lower oxidation potential [21][22] (Figure 5). Reports have revealed that artesunate can cause double-stranded breaks of plasmodial DNA in less than one hour and that it is linked to an increase in ROS generation [23].
Disruption of the conformation of biomolecules by artemisinins (endoperoxides) or other ROS-generating antimalarials kills parasites. Although they have a similar activation process generating carbon- and oxygen-centered radicals, the efficacy of endoperoxides varies, which can be explained, in part, by their different pharmacological properties, notably the stage of the parasite erythrocyte cycle, the nature of their target, and their location in the parasite, i.e., trioxolanes vs. artemisinin and its derivatives [10][24].
3. ROS in Plasmodium-Infected Erythrocytes under Steady State
ROS arise from both the metabolism of the parasite and the host defense system [25]. In erythrocytic parasites, there are two important sources of ROS: the mitochondrial electron transport chain and the degradation of hemoglobin [26][27]. The involvement of the antioxidant machinery and the elimination of heme produced during the digestion of hemoglobin are the two strategies by which the parasite maintains its redox homeostasis (Table 1).Table 1. Plasmodial heme elimination and antioxidant machinery.
| Agent | Site of Production | Role in Oxidative Homeostasis |
|---|---|---|
| HRP | FV | Binding with heme for polymerization |
| HDP | FV | Heme polymerization to hemozoin |
| H2O2 | Cytosol and FV | Degrades heme |
| SOD | Cytosol, Mitochondria | Dismutation of O2•− |
| Prx | Cytosol, Mitochondria, Apicoplast | Reduction of H2O2 to H2O |
| Trx | Cytosol, Mitochondria | Reduction of Prx |
| GSH | Cytosol | Degradation of heme, reduction of proteins and ROS |
| Vit B6 | Cytosol | Role unclear |
H2O2: hydrogen peroxide, HRP: histidine-rich protein, HDP: heme detoxification protein, SOD: superoxide dismutase, Trx: thioredoxin, Prx: peroxiredoxin, GSH: reduced glutathione, Vit B6: vitamin B6, FV: food vacuole [28][29][30][31][32][33][34][35].
4. ROS Production in Plasmodium-Infected Erythrocytes under Antimalarial Treatment
Oxidative stress is not only an important clinical and pathobiochemical factor but also an effective therapeutic principle in malaria. Indeed, the pharmacological activity of some of the most important antimalarials, such as quinolines, atovaquones, and artemisinin derivatives, is mediated by the high production of ROS, exceeding the oxidative stress management capacities of the parasite [36][37].
4.1. Mode of Action of Chloroquine and Other Quinolines
Quinolines have been reported to inhibit the polymerization of heme to hemozoin in food vacuoles, leading to the accumulation of heme [38]. Moreover, chloroquine and other quinolines differently inhibit the peroxidative degradation of heme by H2O2, hence increasing heme accumulation and consequently enhancing heme-catalyzed reactions, leading to the production of ROS and parasite death [29][39]. The formation of the heme–FeIII adduct with chloroquine in the food vacuole leads to its diffusion into the cytosol, where the complex is dissociated, releasing heme in the parasite cytosol and hence promoting the redox cycling of FeIII to FeII, which enhances ROS generation [40]. This has also been reported for quinine [40] and may apply to other quinolines. Other purported modes of action of quinolines include the inhibition of the peroxidase-like activities of heme, leading to the accumulation of H2O2, which interacts with heme to form radicals [41].
4.2. Mode of Action of Atovaquone and Hydroxynaphtoquinones
The mode of action of atovaquone and other hydroxynaphtoquinones is based on the inhibition of mitochondrial cytochrome bc1 complex via the competitive inhibition of ubiquinol binding [42][43]. Consequently, atovaquone induces the collapse of the mitochondrial membrane potential, blocking the energy supply of the parasites, leading to parasite death [44][45]. Concomitantly, the ubiquinol accumulation due to the inhibition of its binding site generates also a significant amount of superoxide radicals (ROS), as was shown in cancer cells and Plasmodium [36][46].
4.3. Mode of Action of Artemisinin: The Role of Endoperoxide
4.3.1. Activation of the Endoperoxide to Generate ROS
The endoperoxides are converted into carbon-centered radicals, which mediate most of their effects. Activation is performed by the transfer of electrons from transition metals, mainly iron (Box 2), leading to the homolytic cleavage of the endoperoxide bridge [47][48][49][50]. The carbon-centered radical formed can alkylate heme, preventing its polymerization into hemozoin and thus inducing the formation of toxic ROS. They also alkylate several essential biomolecules, rendering them dysfunctional and consequently leading to plasmodial death. The iron could be heme (nonchelatable iron) (Figure 9) or freely circulating iron (chelatable iron).4.3.2. Depolarization of the Mitochondrial Membrane Potential
Mitochondria can play an essential role in the indirect killing of the parasite from ROS generation [48][51]. ROS production occurs as a result of membrane depolarization, dissipating its membrane potential, which plays vital roles in maintaining parasite cellular integrity, hence causing death [52][53]. This depolarization can be caused by the direct inhibition of PfATPase6 by artemisinins and other endoperoxides [54]. Artemisinins can also generate ROS at the mitochondrial level by altering the transfer of electrons from complex III to molecular oxygen, forming superoxide radicals [36][23][55][51].4.4. Artemisinin-Based Combination Therapies
As discussed earlier, quinolines and artemisinins act essentially via the production of ROS. The WHO has recommended six artemisinin-based combination therapies (ACTs): artemether–lumefantrine (AL), dihydroartemisinin–piperaquine (DHA-PPQ), artesunate–amodiaquine (AS-AQ), artesunate–mefloquine (AS-MQ), artesunate–sulfadoxine-primethamine (AS-SP), and artesunate–pyronaridine (AS-PY) [56]. ACTs drastically clear the parasite and resolve malaria symptoms (such as fever) [57], hence the need for their continued use. Synthetic hybrids of endoperoxides and quinolines have been developed to enhance their pharmacological activities and cost-effectiveness in malaria treatment [58][59].
4.5. ROS Evasive Mechanisms under Treatment
4.5.1. Preventive Mechanisms
Reduction of self-generated ROS: The parasite is said to reduce its own production of ROS and develop new evolutionary mechanisms to curb the effect of ROS [60]. This evolutionary mechanism undergoes apicoplast metabolism with lipoic acid production, which has antioxidant properties. Lipoic acids are disulfide-containing derivatives of octanoic acid that can exist in oxidized and reduced forms. This ability enables them to play antioxidant roles. Lipoic acids serve as cofactors to several enzymes involved in energy and amino acid metabolism [61]. In the case of artemisinin resistance, reduced hemoglobin import leads to a reduction in the activation of artemisinin and reduced ROS generation [62].
Reduced or mutated expression of hemoglobinases and reduced Hb endocytosis: Endoperoxide activation largely depends on heme production from Hb [63][64]. Downregulation of hemoglobinases (falcipain 2 and 3) in the early ring stage could participate in artemisinin resistance by enhancing the effect of the K13 mutation [64].
4.5.2. Reductive Mechanisms
Import of biomolecules: The host boasts more developed antioxidant machinery against ROS than Plasmodium. Regardless of this, Plasmodium can develop evolutionary measures to eliminate the marauding ROS in its cytosol. Among these measures is the importation of some human antioxidant machinery, such as human peroxiredoxin-2, which uses PfTrx as a reducing agent. Importation is reported to be elevated under treatment with ROS-generating antimalarials [65]. Other forms of human proteins (SOD, catalase, aminolevulinic acid dehydratase, and ferrochelatase) that may augment parasitic defense mechanisms have also been reportedly imported by Plasmodium [66][67][68][69].
Increased expression of antioxidants: Because of the elevated ROS generation due to antimalarial treatment, parasites increase their expression of vital antioxidant enzymes. Such enzymes include iron-superoxide synthetase (Fe-SOD), glutathione-S-transferase (GST), glutathione synthetase (GS), γ-glutamylcysteine synthetase (γ-GCS), thioredoxin reductase (TrxR), and peroxiredoxins (nPrx) [55][70][71]. This overexpression, which is higher in the artemisinins-resistant strains, especially SOD and GST, is associated with parasite resistance characterized by a lower level of ROS and less oxidized proteins [55].
4.5.3. Reparative Mechanisms
ROS generated by antimalarials damage many biomolecules, such as proteins, lipids, and nucleic acids [1]. Increased repair is therefore necessary for continued survival under elevated ROS production. The unfolded protein response (UPR), often referred to as a stress gene, is essential for parasite survival and is upregulated in K13 mutants. This confers upon the parasite an increased ability to repair or degrade proteins damaged by alkylation and oxidation generated by artemisinin [72]. Moreover, K13 mutations have been associated with the elevation of P. falciparum phosphatidylinositol-3-kinase (PfPI3K) due to its reduced association with PfKelch13 and polyubiquitination [73]. This consequently leads to the elevation of phosphatidylinositol-3-phosphate (PI3P), which is said to be essential in the trafficking of proteins and lipids toward the apicoplast, where they are needed [73][74]. K13 gene mutations have demonstrated a global spread and attracted increased attention, especially in endemic regions, as molecular markers for ART resistance [75], although not all have been linked to resistance to artemisinin [76][77]. Single-nucleotide polymorphisms (SNPs) in P. falciparum mlh1, pms1, and exo1 lead to increased expression of thioredoxin (PfTrx) and signal peptide peptidase, which increases adaptation to oxidative stress and protein damage, leading to the upregulation of the DNA repair mechanisms of the parasite with a consequent decrease in the antimalarial effect of artemisinin [71][78]. These mechanisms keep the vital biomolecules of parasites functional and, as a result, improve their chances of survival under treatment, i.e., resistance development.
References
- Santo, A.; Zhu, H.; Li, Y.R. Free Radicals: From Health to Disease. React. Oxyg. Species 2016, 2, 245–263.
- Li, Y.R.; Trush, M. Defining ROS in Biology and Medicine. React. Oxyg. Species 2016, 1, 9–21.
- Hancock, J.T.; Desikan, R.; Neill, S.J. Role of reactive oxygen species in cell signalling pathways. Biochem. Soc. Trans. 2001, 2, 345–350.
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive Oxygen Species in Inflammation and Tissue Injury. Antioxid. Redox Signal. 2014, 20, 1126.
- Stefanatos, R.; Sanz, A. The role of mitochondrial ROS in the aging brain. FEBS Lett. 2018, 592, 743–758.
- Sugamura, K.; Keaney, J.F. Reactive oxygen species in cardiovascular disease. Free Radic. Biol. Med. 2011, 51, 978–992.
- Deavall, D.G.; Martin, E.A.; Horner, J.M.; Roberts, R. Drug-induced oxidative stress and toxicity. J. Toxicol. 2012, 2012, 645460.
- Thannickal, V.J.; Fanburg, B.L. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000, 279, L1005–L1028.
- Sampson, C.; Keens, R.H.; Kattnig, D.R. On the magnetosensitivity of lipid peroxidation: Two- versus three-radical dynamics. Phys. Chem. Chem. Phys. 2019, 21, 13526–13538.
- Hartwig, C.L.; Rosenthal, A.S.; D’Angelo, J.; Griffin, C.E.; Posner, G.H.; Cooper, R.A. Accumulation of artemisinin trioxane derivatives within neutral lipids of Plasmodium falciparum malaria parasites is endoperoxide-dependent. Biochem. Pharmacol. 2009, 77, 322–336.
- Kumura, N.; Furukawa, H.; Onyango, A.N.; Izumi, M.; Nakajima, S.; Ito, H.; Hatano, T.; Kim, H.S.; Wataya, Y.; Baba, N. Different behavior of artemisinin and tetraoxane in the oxidative degradation of phospholipid. Chem. Phys. Lipids 2009, 160, 114–120.
- Fernando, N.; Wickremesinghe, S.; Niloofa, R.; Rodrigo, C.; Karunanayake, L.; De Silva, H.J.; Wickremesinghe, A.R.; Premawansa, S.; Rajapakse, S.; Handunnetti, S.M. Protein carbonyl as a biomarker of oxidative stress in severe leptospirosis, and its usefulness in differentiating leptospirosis from dengue infections. PLoS ONE 2016, 11, e0156085.
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Milzani, A.; Colombo, R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta 2003, 329, 23–38.
- Nyström, T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005, 24, 1311–1317.
- Fedorova, M.; Bollineni, R.C.; Hoffmann, R. Protein carbonylation as a major hallmark of oxidative damage. Mass Spectrom Rev 2014, 33, 79–97.
- Thomas, J.A.; Tan, M.S.Y.; Bisson, C.; Borg, A.; Umrekar, T.R.; Hackett, F.; Hale, V.L.; Vizcay-Barrena, G.; Fleck, R.A.; Snijders, A.P.; et al. A protease cascade regulates release of the human malaria parasite Plasmodium falciparum from host red blood cells. Nat. Microbiol. 2018, 3, 447–455.
- Rosenthal, P.J. Cysteine proteases of malaria parasites. Int. J. Parasitol. 2004, 34, 1489–1499.
- Wu, W.M.; Chen, Y.L.; Zhai, Z.; Xiao, S.H.; Wu, Y.L. Study on the mechanism of action of artemether against schistosomes: The identification of cysteine adducts of both carbon-centred free radicals derived from artemether. Bioorg. Med. Chem. Lett. 2003, 13, 1645–1647.
- Eckstein-Ludwig, U.; Webb, R.J.; Van Goethem, I.D.A.; East, J.M.; Lee, A.G.; Kimura, M.; O’Neill, P.M.; Bray, P.G.; Ward, S.A.; Krishna, S. Artemisinins target the SERCA of Plasmodium falciparum. Nature 2003, 424, 957–961.
- Wang, J.; Xu, C.; Lun, Z.R.; Meshnick, S.R. Unpacking ‘Artemisinin Resistance’. Trends Pharmacol. Sci. 2017, 38, 506–511.
- Jena, N.R. DNA damage by reactive species: Mechanisms, mutation and repair. J. Biosci. 2012, 37, 503–517.
- Cadet, J.; Wagner, J.R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559.
- Gopalakrishnan, A.M.; Kumar, N. Antimalarial action of artesunate involves DNA damage mediated by reactive oxygen species. Antimicrob. Agents Chemother. 2015, 59, 317–325.
- Uhlemann, A.C.; Wittlin, S.; Matile, H.; Bustamante, L.Y.; Krishna, S. Mechanism of antimalarial action of the synthetic trioxolane RBX11160 (OZ277). Antimicrob. Agents Chemother. 2007, 51, 667–672.
- Postma, N.S.; Mommers, E.C.; Eling, W.M.C.; Zuidema, J. Oxidative stress in malaria; implications for prevention and therapy. Pharm. World Sci. 1996, 18, 121–129.
- Rahbari, M.; Rahlfs, S.; Jortzik, E.; Bogeski, I.; Becker, K. H2O2 dynamics in the malaria parasite Plasmodium falciparum. PLoS ONE 2017, 12, e0174837.
- Percário, S.; Moreira, D.R.; Gomes, B.A.Q.; Ferreira, M.E.S.; Gonçalves, A.C.M.; Laurindo, P.S.O.C.; Vilhena, T.C.; Dolabela, M.F.; Green, M.D. Oxidative stress in Malaria. Int. J. Mol. Sci. 2012, 13, 16346–16372.
- Tiwari, S.; Sharma, N.; Sharma, G.P.; Mishra, N. Redox interactome in malaria parasite Plasmodium falciparum. Parasitol. Res. 2021, 120, 423–434.
- Loria, P.; Miller, S.; Foley, M.; Tilley, L. Inhibition of the peroxidative degradation of haem as the basis of action of chloroquine and other quinoline antimalarials. Biochem. J. 1999, 339, 363–370.
- Zhang, J.; Krugliak, M.; Ginsburg, H. The fate of ferriprotorphyrin IX in malaria infected erythrocytes in conjunction with the mode of action of antimalarial drugs. Mol. Biochem. Parasitol. 1999, 99, 129–141.
- Egan, T.J.; Combrinck, J.M.; Egan, J.; Hearne, G.R.; Marques, H.M.; Ntenteni, S.; Sewell, B.T.; Smith, P.J.; Taylor, D.; Van Schalkwyk, D.A.; et al. Fate of haem iron in the malaria parasite Plasmodium falciparum. Biochem. J. 2002, 365, 343–347.
- Rahlfs, S.; Schirmer, R.H.; Becker, K. The thioredoxin system of Plasmodium falciparum and other parasites. Cell. Mol. Life Sci. 2002, 59, 1024–1041.
- Desakorn, V.; Dondorp, A.M.; Silamut, K.; Pongtavornpinyo, W.; Sahassananda, D.; Chotivanich, K.; Pitisuttithum, P.; Smithyman, A.M.; Day, N.P.J.; White, N.J. Stage-dependent production and release of histidine-rich protein 2 by Plasmodium falciparum. Trans. R. Soc. Trop. Med. Hyg. 2005, 99, 517–524.
- Wrenger, C.; Eschbach, M.L.; Müller, I.B.; Warnecke, D.; Walter, R.D. Analysis of the vitamin B6 biosynthesis pathway in the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 2005, 280, 5242–5248.
- Nickel, C.; Rahlfs, S.; Deponte, M.; Koncarevic, S.; Becker, K. Thioredoxin networks in the malarial parasite Plasmodium falciparum. Antioxid. Redox Signal. 2006, 8, 1227–1239.
- Egwu, C.O.; Tsamesidis, I.; Pério, P.; Augereau, J.-M.; Benoit-Vical, F.; Reybier, K. Superoxide: A major role in the mechanism of action of essential antimalarial drugs. Free Radic. Biol. Med. 2021, 167, 271–275.
- Tsamesidis, I.; Egwu, C.O.; Pério, P.; Augereau, J.M.; Benoit-Vical, F.; Reybier, K. An LC–MS assay to measure superoxide radicals and hydrogen peroxide in the blood system. Metabolites 2020, 10, 175.
- Sullivan, D.J.; Matile, H.; Ridley, R.G.; Goldberg, D.E. A common mechanism for blockade of heme polymerization by antimalarial quinolines. J. Biol. Chem. 1998, 273, 31103–31107.
- Sugioka, Y.; Suzuki, M.; Sugioka, K.; Nakano, M. A ferriprotoporphyrin IX-chloroquine complex promotes membrane phospholipid peroxidation A possible mechanism for antimalarial action. FEBS Lett. 1987, 223, 251–254.
- Haynes, R.K.; Cheu, K.W.; Chan, H.W.; Wong, H.N.; Li, K.Y.; Tang, M.M.K.; Chen, M.J.; Guo, Z.F.; Guo, Z.H.; Sinniah, K.; et al. Interactions between Artemisinins and other Antimalarial Drugs in Relation to the Cofactor Model-A Unifying Proposal for Drug Action. ChemMedChem 2012, 7, 2204–2226.
- De Almeida Ribeiro, M.C.; Augusto, O.; Da Costa Ferreira, A.M. Inhibitory effect of chloroquine on the peroxidase activity of ferriprotoporphyrin IX. J. Chem. Soc. Dalt. Trans. 1995, 3759–3766.
- Birth, D.; Kao, W.C.; Hunte, C. Structural analysis of atovaquone-inhibited cytochrome bc 1 complex reveals the molecular basis of antimalarial drug action. Nat. Commun. 2014, 5, 1–11.
- Fry, M.; Pudney, M. Site of action of the antimalarial hydroxynaphthoquinone, 2--3- hydroxy-1,4-naphthoquinone (566C80). Biochem. Pharmacol. 1992, 43, 1545–1553.
- Srivastava, I.K.; Rottenberg, H.; Vaidya, A.B. Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J. Biol. Chem. 1997, 272, 3961–3966.
- Barton, V.; Fisher, N.; Biagini, G.A.; Ward, S.A.; O’Neill, P.M. Inhibiting Plasmodium cytochrome bc1: A complex issue. Curr. Opin. Chem. Biol. 2010, 14, 440–446.
- Fiorillo, M.; Lamb, R.; Tanowitz, H.B.; Mutti, L.; Krstic-Demonacos, M.; Cappello, A.R.; Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Repurposing atovaquone: Targeting mitochondrial complex III and OXPHOS to eradicate cancer stem cells. Oncotarget 2016, 7, 34084–34099.
- Posner, G.H.; Oh, C.H. A Regiospecifically Oxygen-18 Labeled 1,2,4-Trioxane: A Simple Chemical Model System To Probe the Mechanism(s) for the Antimalarial Activity of Artemisinin (Qinghaosu). J. Am. Chem. Soc. 1992, 114, 8328–8329.
- Mercer, A.E.; Copple, I.M.; Maggs, J.L.; O’Neill, P.M.; Park, B.K. The role of heme and the mitochondrion in the chemical and molecular mechanisms of mammalian cell death induced by the artemisinin antimalarials. J. Biol. Chem. 2011, 286, 987–996.
- Posner, G.H.; Oh, C.H.; Wang, D.; Gerena, L.; Milhous, W.K.; Meshnick, S.R.; Asawamahasadka, W. Mechanism-Based Design, Synthesis, and in Vitro Antimalarial Testing of New 4-Methylated Trioxanes Structurally Related to Artemisinin: The Importance of a Carbon-Centered Radical for Antimalarial Activity. J. Med. Chem. 1994, 37, 1256–1258.
- Robert, A.; Benoit-Vical, F.; Claparols, C.; Meunier, B. The antimalarial drug artemisinin alkylates heme in infected mice. Proc. Natl. Acad. Sci. USA 2005, 102, 13676–13680.
- Wang, J.; Huang, L.; Li, J.; Fan, Q.; Long, Y.; Li, Y.; Zhou, B. Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS ONE 2010, 5, e9582.
- Allen, R.J.W.; Kirk, K. The Membrane Potential of the Intraerythrocytic Malaria Parasite Plasmodium falciparum. J. Biol. Chem. 2004, 279, 11264–11272.
- Biagini, G.A.; Viriyavejakul, P.; O’Neill, P.M.; Bray, P.G.; Ward, S.A. Functional characterization and target validation of alternative complex I of Plasmodium falciparum mitochondria. Antimicrob. Agents Chemother. 2006, 50, 1841–1851.
- Antoine, T.; Fisher, N.; Amewu, R.; O’Neill, P.M.; Ward, S.A.; Biagini, G.A. Rapid kill of malaria parasites by artemisinin and semi-synthetic endoperoxides involves ROS-dependent depolarization of the membrane potential. J. Antimicrob. Chemother. 2014, 69, 1005–1016.
- Egwu, C.O.; Pério, P.; Augereau, J.-M.; Tsamesidis, I.; Benoit-Vical, F.; Reybier, K. Resistance to artemisinin in falciparum malaria parasites: A redox-mediated phenomenon. Free Radic. Biol. Med. 2021, S0891-5849, 00476–00477.
- WHO. Artemisinin Resistance and Artemisinin-Based Combination Therapy Efficacy (Status Report—August 2018). Available online: https://apps.who.int/iris/bitstream/handle/10665/274362/WHO-CDS-GMP-2018.18-eng.pdf?ua=1 (accessed on 26 December 2020).
- Van Vugt, M.; Brockman, A.; Gemperli, B.; Luxemburger, C.; Gathmann, I.; Royce, C.; Slight, T.; Looareesuwan, S.; White, N.J.; Nosten, F. Randomized comparison of artemether-benflumetol and artesunate-mefloquine in treatment of multidrug-resistant falciparum malaria. Antimicrob. Agents Chemother. 1998, 42, 135–139.
- Rudrapal, M.; Chetia, D. Endoperoxide antimalarials: Development, structural diversity and pharmacodynamic aspects with reference to 1,2,4-trioxane-based structural scaffold. Drug Des. Devel. Ther. 2016, 10, 3575–3590.
- Benoit-Vical, F.; Lelièvre, J.; Berry, A.; Deymier, C.; Dechy-Cabaret, O.; Cazelles, J.; Loup, C.; Robert, A.; Magnaval, J.F.; Meunier, B. Trioxaquines are new antimalarial agents active on all erythrocytic forms, including gametocytes. Antimicrob. Agents Chemother. 2007, 51, 1463–1472.
- Toler, S. The plasmodial apicoplast was retained under evolutionary selective pressure to assuage blood stage oxidative stress. Med. Hypotheses 2005, 65, 683–690.
- Storm, J. Lipoic Acid Metabolism of Plasmodium—A Suitable Drug Target. Curr. Pharm. Des. 2012, 18, 3480.
- Birnbaum, J.; Scharf, S.; Schmidt, S.; Jonscher, E.; Maria Hoeijmakers, W.A.; Flemming, S.; Toenhake, C.G.; Schmitt, M.; Sabitzki, R.; Bergmann, B.; et al. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science 2020, 367, 51–59.
- Zhang, S.; Gerhard, G.S. Heme activates artemisinin more efficiently than hemin, inorganic iron, or hemoglobin. Bioorg. Med. Chem. 2008, 16, 7853–7861.
- Xie, S.C.; Dogovski, C.; Hanssen, E.; Chiu, F.; Yang, T.; Crespo, M.P.; Stafford, C.; Batinovic, S.; Teguh, S.; Charman, S.; et al. Haemoglobin degradation underpins the sensitivity of early ring stage Plasmodium falciparum to artemisinins. J. Cell Sci. 2016, 129, 406–416.
- Koncarevic, S.; Rohrbach, P.; Deponte, M.; Krohne, G.; Prieto, J.H.; Yates, J.; Rahlfs, S.; Becker, K. The malarial parasite Plasmodium falciparum imports the human protein peroxiredoxin 2 for peroxide detoxification. Proc. Natl. Acad. Sci. USA 2009, 106, 13323–13328.
- Fairfield, A.S.; Meshnick, S.R.; Eaton, J.W. Malaria parasites adopt host cell superoxide dismutase. Science 1983, 221, 764–766.
- Clarebout, G.; Slomianny, C.; Delcourt, P.; Leu, B.; Masset, A.; Camus, D.; Dive, D. Status of Plasmodium falciparum towards catalase. Br. J. Haematol. 1998, 103, 52–59.
- Varadharajan, S.; Sagar, B.K.C.; Rangarajan, P.N.; Padmanaban, G. Localization of ferrochelatase in Plasmodium falciparum. Biochem. J. 2004, 384, 429–436.
- Bonday, Z.Q.; Dhanasekaran, S.; Rangarajan, P.N.; Padmanaban, G. Import of host δ-aminolevulinate dehydratase into the malarial parasite: Identification of a new drug target. Nat. Med. 2000, 6, 898–903.
- Nogueira, F.; Diez, A.; Radfar, A.; Pérez-Benavente, S.; Rosario, V.E.; Puyet, A.; Bautista, J.M. Early transcriptional response to chloroquine of the Plasmodium falciparum antioxidant defence in sensitive and resistant clones. Acta Trop. 2010, 114, 109–115.
- Rocamora, F.; Zhu, L.; Liong, K.Y.; Dondorp, A.; Miotto, O.; Mok, S.; Bozdech, Z. Oxidative stress and protein damage responses mediate artemisinin resistance in malaria parasites. PLoS Pathog. 2018, 14, e1006930.
- Mok, S.; Ashley, E.A.; Ferreira, P.E.; Zhu, L.; Lin, Z.; Yeo, T.; Chotivanich, K.; Imwong, M.; Pukrittayakamee, S.; Dhorda, M.; et al. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 2015, 347, 431–435.
- Mbengue, A.; Bhattacharjee, S.; Pandharkar, T.; Liu, H.; Estiu, G.; Stahelin, R.V.; Rizk, S.S.; Njimoh, D.L.; Ryan, Y.; Chotivanich, K.; et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 2015, 520, 683–687.
- Tawk, L.; Chicanne, G.; Dubremetz, J.F.; Richard, V.; Payrastre, B.; Vial, H.J.; Roy, C.; Wengelnik, K. Phosphatidylinositol 3-phosphate, an essential lipid in Plasmodium, localizes to the food vacuole membrane and the apicoplast. Eukaryot. Cell 2010, 9, 1519–1530.
- Ariey, F.; Witkowski, B.; Amaratunga, C.; Beghain, J.; Langlois, A.C.; Khim, N.; Kim, S.; Duru, V.; Bouchier, C.; Ma, L.; et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014, 505, 50–55.
- Isozumi, R.; Uemura, H.; Kimata, I.; Ichinose, Y.; Logedi, J.; Omar, A.H.; Kaneko, A. Novel mutations in k13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg. Infect. Dis. 2015, 21, 490–492.
- Balikagala, B.; Mita, T.; Ikeda, M.; Sakurai, M.; Yatsushiro, S.; Takahashi, N.; Tachibana, S.I.; Auma, M.; Ntege, E.H.; Ito, D.; et al. Absence of in vivo selection for K13 mutations after artemether-lumefantrine treatment in Uganda. Malar. J. 2017, 16, 23.
- Lee, A.H.; Fidock, D.A. Evidence of a mild mutator phenotype in cambodian Plasmodium falciparum malaria parasites. PLoS ONE 2016, 11, e0154166.
More
