Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Agnieszka Fiszer.
During polyadenylation, a polyadenosine sequence-namely, a poly(A) tail-is added to the 3′ end of a transcript. Together with the removal of introns and the addition of a 5′ cap, polyadenylation constitutes a major step in pre-mRNA maturation. The polyadenylation process can be divided into two major steps: first, newly transcribed pre-mRNA is cleaved and its 3′ end is generated; then, a specific enzyme-poly(A) polymerase (PAP)-generates the poly(A) tail independently from the template, starting from the cleavage site.
- polyglutamine diseases
- Huntington’s disease
- poly(A) tail
- alternative polyadenylation
- repeat expansion diseases
1. Polyadenylation Process
During polyadenylation, a polyadenosine sequence-namely, a poly(A) tail-is added to the 3′ end of a transcript. Together with the removal of introns and the addition of a 5′ cap, polyadenylation constitutes a major step in pre-mRNA maturation [1,3,4][1][2][3]. The polyadenylation process can be divided into two major steps: first, newly transcribed pre-mRNA is cleaved and its 3′ end is generated; then, a specific enzyme-poly(A) polymerase (PAP)-generates the poly(A) tail independently from the template, starting from the cleavage site [5,6,7][4][5][6]. These two steps depend on the interplay between various cis- and trans-acting factors (Figure 1). Cis elements, which are typically located in 3′-UTRs, determine the position of cleavage and polyadenylation site in mRNA [8][7]. The most crucial cis-element is an RNA sequence motif called the polyadenylation signal (PAS). In animals, the canonical PAS is the hexanucleotide sequence AAUAAA [9,10,11][8][9][10]. The cleavage site is located around 10 to 30 nucleotides downstream from the PAS, between the PAS and a core downstream sequence element (DSE)-a U-/GU-rich sequence, which is itself located 14–70 nucleotides downstream of the cleavage site (Figure 1) [12][11]. The cleavage itself occurs just before an adenosine residue, mostly after cytosine. Another cis-element that is involved in the polyadenylation process is a U-rich/UGUA upstream sequence element (USE) located upstream of PAS. All three cis-elements are recognized by their respective trans-acting factors, which interact with each other through the carboxyl-terminal domain (CTD) of RNA polymerase II (RNAPII) [5][4]. PAS recruits cleavage and polyadenylation specificity factor (CPSF), DSE is recognized by cleavage stimulation factor (CstF), and USE recruits cleavage factor Im (CFIm). Cooperation between those factors leads to cleavage. The subsequent cooperation between nuclear poly(A)-binding protein (PABPN1) and PAP allows for the generation of the poly(A) tail at the cleavage site: PABPN1 acts as a type of ‘ruler’, which is important for the synthesis of an appropriately sized poly(A) tail, while PAP performs a non-templated addition of adenosine residues [13][12].
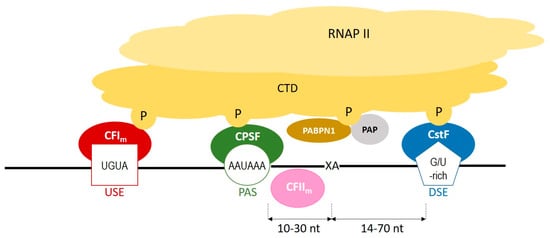
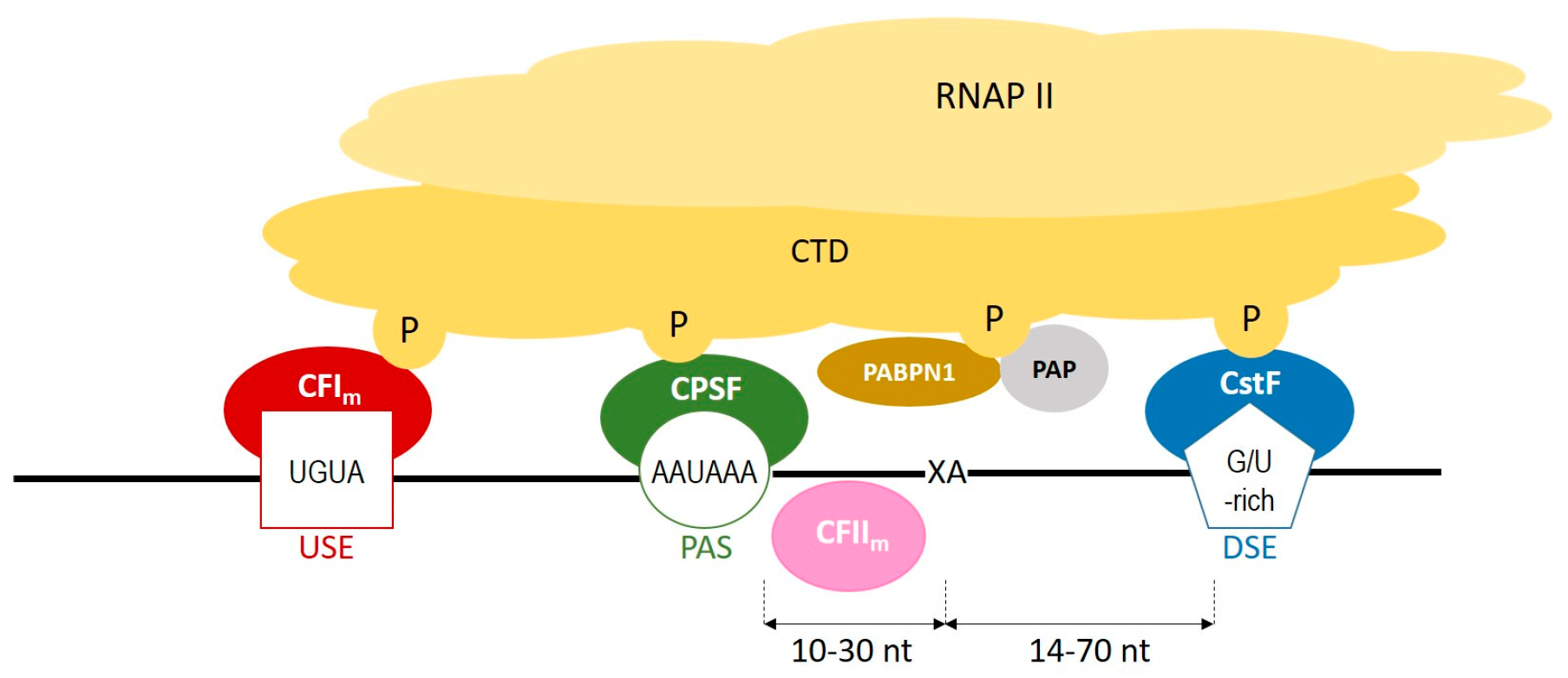 Figure 1. A scheme representing the cooperation between cis-elements and trans-factors, which are involved in a polyadenylation process. CFIm–cleavage factor Im; CFIIm–cleavage factor IIm; CPSF–cleavage and polyadenylation specificity factor; CstF–cleavage stimulation factor; CTD–carboxyterminal domain of RNA polymerase II; DSE–downstream element; P–phosphorylated serines of CTD; PABPN1–nuclear poly(A) binding protein; PAP–poly(A) polymerase; PAS–poly(A) signal; RNAPII–RNA polymerase II; USE–upstream element; XA–cleavage site.
Figure 1. A scheme representing the cooperation between cis-elements and trans-factors, which are involved in a polyadenylation process. CFIm–cleavage factor Im; CFIIm–cleavage factor IIm; CPSF–cleavage and polyadenylation specificity factor; CstF–cleavage stimulation factor; CTD–carboxyterminal domain of RNA polymerase II; DSE–downstream element; P–phosphorylated serines of CTD; PABPN1–nuclear poly(A) binding protein; PAP–poly(A) polymerase; PAS–poly(A) signal; RNAPII–RNA polymerase II; USE–upstream element; XA–cleavage site.2. Polyadenylation Roles
The poly(A) tail plays an important role in a transcript’s life cycle [2][13]. For example, it is responsible for mediating the transport of mature mRNA into the cytoplasm through the NXF1-dependent pathway using nuclear-pore complexes embedded in the nuclear envelope [14]. Poly(A) tails are also involved in maintaining the stability of transcripts. They participate in the regulation of mRNA degradation through the process of deadenylation, which is commonly activated in post-transcriptional regulation via the sequence-specific binding of microRNA (miRNA), together with miRNA-induced silencing complex (miRISC), to the 3′-UTR [15,16][15][16]. Poly(A) tails are also important for translation regulation [17,18][17][18]. It is suggested that they interact functionally and physically with the 5′ cap of mRNA, creating a closed-loop structure that promotes the initiation of translation.
In metazoa, almost all mRNAs undergo polyadenylation. One of the exceptions is replication-dependent histone protein mRNAs, which form a highly conserved stem-loop structure [19,20][19][20]. Additionally, according to a recent comprehensive study [21], nearly 50% of long non-coding RNAs (lncRNAs) undergo polyadenylation, and the resulting poly(A) tails are important for their regulation at the cellular level [22].
3. Length and Composition of Tails
The lengths of poly(A) tails can vary between transcripts. For example, mRNAs of highly expressed genes, such as housekeeping genes, usually possess shorter poly(A) tails, whereas poorly translated transcripts and lncRNAs have longer tails [23,24][23][24]. Generally, the length range for specifying a short or long tail is dependent on the population of transcripts analyzed. For example, in a Caenorhabditis elegans study, half of the transcripts possessed poly(A) tails in the range of 70 to 94 A residues; therefore, short tails were defined as those with ≤70 A residues, and long ones, as those with >94 A residues [23]. The existence of shorter tails may be also explained by the fact that some genes (e.g., albumin and transferrin) contain poly(A)-limiting element (PLE), which tends to restrict the initial length of poly(A) tails from the pre-mRNA stage [25,26][25][26]. Additionally, a cytoplasmic polyadenylation element (CPE) can modulate the length of a poly(A) tail after the export of the transcript from the nucleus [27]. It is located in the 3′-UTR, near PAS, and its most common sequence is UUUUAU [28]. As investigated in neurons and during early development, CPE can influence the length of the poly(A) tail through the binding of proteins to this motif (e.g., CPEB) [29]. CPEB can promote either cytoplasmic polyadenylation or deadenylation. When it is deactivated (dephosphorylated), it recruits poly(A) ribonuclease deadenylase (PARN) to deadenylate and repress the expression of mRNA. On the other hand, when it is phosphorylated, it promotes the expulsion of PARN, thereby leading to polyadenylation by germ-line development factor 2 (GLD2) [26,30][26][30]. This cytoplasmic polyadenylation process occurs in mRNAs that already contain a short poly(A) tail and usually activates translation, leading to increased protein expression.
An interesting fact is that poly(A) tails are not limited to possessing A residues: reports indicate that cytosines, guanosines, and uridines can also be incorporated into the poly(A) tail. While the role of cytosines added to the poly(A) tail remains to be elucidated, the guanylation and uridylation of poly(A) tails are quite well understood. Guanylation occurs only for longer poly(A) tails and can slow down deadenylation, delaying transcript decay [31]. Guanylation frequently occurs for long-lived transcripts with a slow turnover, e.g., transcripts encoding secreted proteins [31]. On the other hand, uridylation is usually found in short tails and marks transcripts for decay [32,33][32][33]. This process tends to be a key factor in germline development, differentiation, and early embryogenesis, where short-lived transcripts, with relatively fast turnover, predominate [33]. The lengths of poly(A) tails might also depend on circadian rhythms and the cell cycle [34,35,36][34][35][36]. By studying multiple mouse liver mRNAs, researchers demonstrated that rhythmic changes in poly(A) tail lengths were under the control of the circadian clock. Even more importantly, they presented data indicating that rhythmic poly(A) tail lengths are correlated with rhythmic protein expression [34]. With regard to the cell cycle, TAIL-Seq analysis suggests that global RNA decay takes place during the S phase through the accumulation of terminal uridylation. On the other hand, the accumulation of terminal guanylation occurs during the M phase of the cell cycle, leading to the assumption that the majority of the transcriptome is then protected from active deadenylation [35].
4. Alternative Polyadenylation (APA)
Apart from the canonical PAS-the AAUAAA hexamer-other weaker signals called alternative PASs, may be present in transcripts. Generally, the higher the sequence similarity between an alternative and canonical PAS, the stronger the recognition of the alternative PAS. When this alternative PAS is selected as a signal for the cleavage and polyadenylation event, the process is described as alternative polyadenylation (APA). APA is thought to occur for around 70% of human protein-coding genes and can also affect non-coding RNAs, such as lncRNAs [37,38,39][37][38][39]. The affected transcripts can exhibit various numbers of APA events in a few or multiple APA sites. APA can dramatically modulate the expression of a specific gene and affect the fate of its transcript, including its half-life and cellular localization [40,41][40][41]. Depending on the alternative PAS localization, APA can occur either in 3′-UTRs (UTR-APA) or upstream of the last exon: in introns or protein-coding exons (UR-APA). UR-APA can lead to the production of truncated proteins with different functions (protein diversification), or the production of dysfunctional proteins. On the other hand, when APA occurs in the 3′-UTR of a transcript, it leads to the creation of an mRNA of different lengths, which still codes for a full-length protein. In such cases, APA can affect the expression of a gene by, for example, changing the number of miRNA-binding sites in the transcript. As it was shown that the 3′-UTR can regulate protein localization independently from mRNA localization, it can act as a scaffold for various protein complexes which, when recruited to translation sites, can interact with specific domains of newly translated proteins [42]. This, in turn, leads to the translocation of such proteins. An example is a CD47 transcript, whose short 3′-UTR promotes the localization of the protein at the ER, while its longer isoform promotes its translocation to the plasma membrane [37]. The occurrence of APA can be regulated in many ways, one being the ‘strength’ of alternative PASs. The more similar the sequence of an alternative PAS is to that of the canonical PAS, the stronger the alternative PAS will be. Moreover, the localization of a specific PAS within a transcript sequence is also worth mentioning. Typically, PASs localized closer to the start codon (proximal) are considered to be weaker, while PASs localized further from the start codon (distal) are stronger [5][4]. Core polyadenylation factors, as well as other RNA-binding proteins (RBPs), can also regulate APA. For example, PABPN1 enhances the selection of distal PASs by competing with cleavage and polyadenylation complexes: it recognizes a weak PAS and binds to it, thereby blocking CPSF binding [43,44][43][44]. Regarding RBPs, HuR protein favors the selection of a distal PAS by binding to U-rich elements lying close to a proximal PAS [45]. Another crucial group of RBPs involved in APA is muscleblind-like (MBNL) proteins, whose binding sites are present in the close vicinity of many PASs. In myotonic dystrophy (DM), MBNL proteins were shown to either activate or suppress polyadenylation at specific sites [46]. PABPN1, MBNL, and HuR proteins are described in more detail in the following chapters.
References
- Casañal, A.; Kumar, A.; Hill, C.H.; Easter, A.D.; Emsley, P.; Degliesposti, G.; Gordiyenko, Y.; Santhanam, B.; Wolf, J.; Wiederhold, K.; et al. Architecture of eukaryotic mRNA 3′-end processing machinery. Science 2017, 358, 1056–1059.
- Nourse, J.; Spada, S.; Danckwardt, S. Emerging roles of RNA 3′-end cleavage and polyadenylation in pathogenesis, diagnosis and therapy of human disorders. Biomolecules 2020, 10, 915.
- Stewart, M. Polyadenylation and nuclear export of mRNAs. J. Biol. Chem. 2019, 294, 2977–2987.
- Di Giammartino, D.C.; Nishida, K.; Manley, J.L. Mechanisms and Consequences of Alternative Polyadenylation. Mol. Cell 2011, 43, 853–866.
- Sadek, J.; Omer, A.; Hall, D.; Ashour, K.; Gallouzi, I.E. Alternative polyadenylation and the stress response. Wiley Interdiscip. Rev. RNA 2019, 10, 1–16.
- Ren, F.; Zhang, N.; Zhang, L.; Miller, E.; Pu, J.J. Alternative Polyadenylation: A new frontier in post transcriptional regulation. Biomark. Res. 2020, 8, 67.
- Danckwardt, S.; Hentze, M.W.; Kulozik, A.E. 3′ end mRNA processing: Molecular mechanisms and implications for health and disease. EMBO J. 2008, 27, 482–498.
- Proudfoot, N.J. Ending the message: Poly(A) signals then and now. Genes Dev. 2011, 25, 1770–1782.
- Derti, A.; Garrett-Engele, P.; MacIsaac, K.D.; Stevens, R.C.; Sriram, S.; Chen, R.; Rohl, C.A.; Johnson, J.M.; Babak, T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012, 22, 1173–1183.
- Gruber, A.J.; Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 2019, 20, 599–614.
- Zhao, J.; Hyman, L.; Moore, C. Formation of mRNA 3′ Ends in Eukaryotes: Mechanism, Regulation, and Interrelationships with Other Steps in mRNA Synthesis. Microbiol. Mol. Biol. Rev. 1999, 63, 405–445.
- Eckmann, C.R.; Rammelt, C.; Wahle, E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA 2011, 2, 348–361.
- Passmore, L.A.; Coller, J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat. Rev. Mol. Cell Biol. 2021.
- Natalizio, B.J.; Wente, S.R. Postage for the messenger: Designating routes for nuclear mRNA export. Trends Cell Biol. 2013, 23, 365–373.
- Bresson, S.M.; Conrad, N.K. The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay. PLoS Genet. 2013, 9, 1003893.
- Eisen, T.J.; Eichhorn, S.W.; Subtelny, A.O.; Lin, K.S.; McGeary, S.E.; Gupta, S.; Bartel, D.P. The Dynamics of Cytoplasmic mRNA Metabolism. Mol. Cell 2020, 77, 786–799.
- Weill, L.; Belloc, E.; Bava, F.-A.; Méndez, R. Translational control by changes in poly(A) tail length: Recycling mRNAs. Nat. Struct. Mol. Biol. 2012, 19, 577–585.
- Xiang, K.; Bartel, D.P. The molecular basis of coupling between poly(A)-tail length and translational efficiency. eLife 2021, 10, 66493.
- Nicholson, A.L.; Pasquinelli, A.E. Tales of Detailed Poly(A) Tails. Trends Cell Biol. 2019, 29, 191–200.
- Dominski, Z.; Yang, X.-C.; Kaygun, H.; Dadlez, M.; Marzluff, W.F. A 3′ Exonuclease that Specifically Interacts with the 3′ End of Histone mRNA. Mol. Cell 2003, 12, 295–305.
- Lorenzi, L.; Chiu, H.S.; Avila Cobos, F.; Gross, S.; Volders, P.J.; Cannoodt, R.; Nuytens, J.; Vanderheyden, K.; Anckaert, J.; Lefever, S.; et al. The RNA Atlas expands the catalog of human non-coding RNAs. Nat. Biotechnol. 2021, 39, 1453–1465.
- Chen, L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772.
- Lima, S.A.; Chipman, L.B.; Nicholson, A.L.; Chen, Y.H.; Yee, B.A.; Yeo, G.W.; Coller, J.; Pasquinelli, A.E. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 2017, 24, 1057–1063.
- Subtelny, A.O.; Eichhorn, S.W.; Chen, G.R.; Sive, H.; Bartel, D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 2014, 508, 66–71.
- Gu, H.; Gupta, J.D.; Schoenberg, D.R. The poly(A)-limiting element is a conserved cis-acting sequence that regulates poly(A) tail length on nuclear pre-mRNAs. Proc. Natl. Acad. Sci. USA 1999, 96, 8943–8948.
- Jalkanen, A.L.; Coleman, S.J.; Wilusz, J. Determinants and implications of mRNA poly(A) tail size—Does this protein make my tail look big? Semin. Cell Dev. Biol. 2014, 34, 24–32.
- Charlesworth, A.; Meijer, H.A.; de Moor, C.H. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip. Rev. RNA 2013, 4, 437.
- Ivshina, M.; Lasko, P.; Richter, J.D. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 2014, 30, 393–415.
- Mendez, R.; Richter, J.D. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2001, 2, 521–529.
- Udagawa, T.; Swanger, S.A.; Takeuchi, K.; Kim, J.H.; Nalavadi, V.; Shin, J.; Lorenz, L.J.; Zukin, R.S.; Bassell, G.J.; Richter, J.D. Bidirectional Control of mRNA Translation and Synaptic Plasticity by the Cytoplasmic Polyadenylation Complex. Mol. Cell 2012, 47, 253–266.
- Lim, J.; Kim, D.; Lee, Y.S.; Ha, M.; Lee, M.; Yeo, J.; Chang, H.; Song, J.; Ahn, K.; Kim, V.N. Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science 2018, 361, 701–704.
- Lim, J.; Ha, M.; Chang, H.; Kwon, S.C.; Simanshu, D.K.; Patel, D.J.; Kim, V.N. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 2014, 159, 1365–1376.
- Morgan, M.; Much, C.; DiGiacomo, M.; Azzi, C.; Ivanova, I.; Vitsios, D.M.; Pistolic, J.; Collier, P.; Moreira, P.N.; Benes, V.; et al. MRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 2017, 548, 347–351.
- Kojima, S.; Sher-Chen, E.L.; Green, C.B. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012, 26, 2724–2736.
- Liu, Y.; Nie, H.; Lu, F. Dynamic RNA 3′ Uridylation and Guanylation during Mitosis. iScience 2020, 23.
- Park, J.E.; Yi, H.; Kim, Y.; Chang, H.; Kim, V.N. Regulation of Poly(A) Tail and Translation during the Somatic Cell Cycle. Mol. Cell 2016, 62, 462–471.
- Tian, B.; Manley, J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2016, 18, 18–30.
- Sommerkamp, P.; Cabezas-Wallscheid, N.; Trumpp, A. Alternative Polyadenylation in Stem Cell Self-Renewal and Differentiation. Trends Mol. Med. 2021, 27, 660–672.
- Elkon, R.; Ugalde, A.P.; Agami, R. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat. Rev. Genet. 2013, 14, 496–506.
- Lutz, C.S.; Moreira, A. Alternative mRNA polyadenylation in eukaryotes: An effective regulator of gene expression. Wiley Interdiscip. Rev. RNA 2011, 2, 22–31.
- Agarwal, V.; Lopez-Darwin, S.; Kelley, D.R.; Shendure, J. The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nat. Commun. 2021, 12, 5101.
- Berkovits, B.D.; Mayr, C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 2015, 522, 363–367.
- Jenal, M.; Elkon, R.; Loayza-Puch, F.; Van Haaften, G.; Kühn, U.; Menzies, F.M.; Vrielink, J.A.F.O.; Bos, A.J.; Drost, J.; Rooijers, K.; et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 2012, 149, 538–553.
- Batra, R.; Manchanda, M.; Swanson, M.S. Global insights into alternative polyadenylation regulation. RNA Biol. 2015, 12, 597–602.
- Dai, W.; Zhang, G.; Makeyev, E.V. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res. 2012, 40, 787–800.
- Batra, R.; Charizanis, K.; Manchanda, M.; Mohan, A.; Li, M.; Finn, D.J.; Goodwin, M.; Zhang, C.; Sobczak, K.; Thornton, C.A.; et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell 2014, 56, 311–322.
More
