Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Jie Zhang and Version 2 by Catherine Yang.
The histopathological image is widely considered as the gold standard for the diagnosis and prognosis of human cancers. Recently, deep learning technology has been extremely successful in the field of computer vision, which has also boosted considerable interest in digital pathology analysis. Deep learning and its extensions have opened several avenues to tackle many challenging histopathological image analysis problems including color normalization, image segmentation, and the diagnosis/prognosis of human cancers.
- machine learning
- digital pathology image analysis
1. Pathology Image Segmentation
The segmentation task, which aims at assigning a class label to each pixel of an image, is a common task in pathology image analysis [1]. The segmentation task on histopathological images can be divided into two categories, nuclei segmentation, and tissue segmentation. The nuclei segmentation task focuses on exploring the nuclei features, such as morphological appearances in histopathological images, which are widely recognized as the most frequently used biomarkers for cancer histology diagnosis. On the other hand, the tissue segmentation task takes the histopathology image as input and segments the tissues that are composed of a group of cells in the input image with certain characteristics and structures (i.e., gland, tumor-infiltrating lymphocytes, etc.). These quantitatively measured tissues are also a crucial indicator for the diagnosis and prognosis of human cancers [2][3].
Due to the heterogenous patterns in WSI, the accurate segmentation of nuclei and tissues in the histopathological images is with huge challenges. First, there are variations on nucleus/tissue sizes and shape, requiring a segmentation model with a strong generalization ability. Second, nuclei/cells are often clustered into clumps so that they might partially overlap or touch one another, which will lead to the under-segmentation of histopathological images. Third, in some malignant cases, such as moderately and poorly differentially adenocarcinomas, the structure of the tissues (such as the glands) are heavily degenerated, making them difficult to discriminate [4][5].
In view of these challenges, numerous deep learning-based approaches have been proposed to extract high-level features from WSI that can achieve enhanced segmentation performance. Here, the researcherswe first review the deep learning-based nuclei segmentation algorithm. Then, the researchers swe summarize the development of deep learning algorithms on tissue-level segmentation tasks. The researchers shWe show the overview of papers using deep learning for nuclei/tissue segmentation in Figure 1.
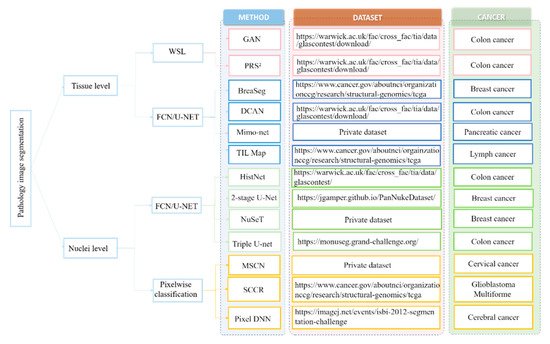
1.1. Nuclei-Level Segmentation
Cellular object segmentation is a prerequisite step for the assessment of human cancers [18]. For example, the counting of mitoses is one of the most prognostic factors in breast cancer requiring the assistance of nuclei segmentation [19]. In the diagnosis of cervical cytology, nuclei segmentation is necessary to discover all types of cytological abnormalities [20]. The traditional nuclei segmentation algorithms are based on morphological processing methods [21], clustering algorithms [22], level set methods [23], and their variants [24][25][26], whose performance are largely determined by the designed features requiring the domain knowledge of experts. Recently, deep learning approaches have been widely applied without the efforts of designing hand-crafted features [27].
Generally, the deep learning-based nuclei segmentation algorithms can be divided into two categories, the pixel-wise classification methods [17][28][29][30] and the fully convolutional network (FCN)-based methods [13][14][31]. Pixel-wise classification methods convert the segmentation task into the classification task, by which the label of each pixel is predicted from raw pixel values in a square window centered on it [28]. For example, Cireşan et al. [17] first densely sampled the squared windows from the WSI, followed by classifying the centered pixels via utilizing the rich context information within the sampled windows. Moreover, Zhou et al. [16] learned a bank of convolutional filters and a sparse linear regressor to produce the likelihood for each pixel being nuclear or background regions. By considering the windows of different sizes can extract helpful complementary information for the nuclei segmentation, a multiscale convolutional network and graph-partitioning–based method [15] were proposed for the task of nuclei segmentation. In addition, Xing et al. [32] firstly learned a CNN model to generate a probability map of each image. According to the probability map, each pixel is then assigned a probability belonging to the nucleus. Finally, an iterative region merging algorithm was used to accomplish the segmentation task. Nesma et al. [33] also presented an optimized pixel-based classification model by the cooperation of region growing strategy that could successfully obtain nucleus and cytoplasm segmentation results. Additionally, Liu et al. [29] proposed a panoptic segmentation model which incorporates an auxiliary semantic segmentation branch with the instance branch to integrate global and local features for nuclei segmentation.
Although the above pixel-wise classification methods have shown more promising performance over the traditional segmentation algorithms, obvious limitations can also be found. First, they are quite slow since the densely selected patches increase the calculation burden for neural network training [34]. Second, the extracted patches cannot fully reveal the rich context information within the whole input image for nuclei segmentation. Accordingly, a more elegant architecture called “fully convolutional network” is proposed [35]. FCN can use the full image rather than the densely extracted patches as the input, which can produce a more accurate and efficient nuclei segmentation result. In addition to FCN, U-Net is another powerful nuclei segmentation tool [36]. In comparison with FCN, U-Net uses skip connections between downsampling and upsampling paths that can stabilize gradient updates for deep model training. Based on the U-Net structure, Zhao et al. [14] proposed a Triple U-Net architecture for nuclei segmentation without the necessity of color normalization and achieved state-of-the-art nuclei segmentation performance (Figure 2). To split touching nuclei that are hard to segment, Yang et al. [13] used a hybrid network consisting of U-Net and region proposal networks, followed by a watershed step to separate them into individual ones. Amirreza et al. [12] proposed a two-stage U-Net–based model for touching cell segmentation, where the first stage used the U-Net to separate nuclei from the background while the second stage applied the U-Net to regress the distance map of each nucleus for the final touching cell segmentation. To explicitly mimic how human pathologists combine multi-scale information, Schmitz et al. [31] introduced a family of multi-encoder FCN with deep fusion for nuclei segmentation. Other U-Net–based studies include [4][11] proposed deep contour-aware networks that integrate multilevel contextual features to accurately detect and segment nuclei from histopathological images, which could also effectively improve the final segmentation performance.
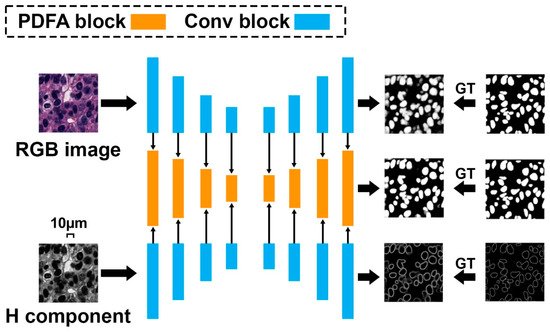
Figure 2. Triple U-Net: hematoxylin-aware nuclei segmentation with progressive dense feature aggregation.
1.2. Tissue-Level Segmentation
Besides nuclei segmentation, computerized segmentation of specific tissues in histopathological images is another core operation to study the tumor biology system. For instance, the segmentation of tumor-infiltrating lymphocytes and characterizing their spatial correlation on WSI have become crucial in diagnosis, prognosis, and treatment response prediction for different cancers [37]. Moreover, gland segmentation is one prerequisite step for quantitatively measuring glandular formation, which is also an important indicator for exploring the degree of differentiation [38][39].
The automatic segmentation of tissues in histology images has been explored by many studies [40][41]. Traditional tissue segmentation methods usually relied on the extraction of handcrafted features, the design of conventional classifiers [42]. Recently, deep learning has become popular in computer vision and image-processing tasks due to its outstanding performance, and some studies also applied deep learning methods for the segmentation of different types of tissues from WSI [9][43][44]. Among the existing deep learning segmentation algorithms, the U-Net-based neural network is still most widely used. For example, Saltz et al. [10] applied the U-Net network to present mappings of tumor-infiltrating lymphocytes on H&E images from 13 TCGA (The Cancer Genome Atlas) tumor types. Based on U-Net, Raza et al. [9] presented a minimal information loss dilated network for gland instance segmentation in colon histology images. Chen et al. [43] presented a deep contour-aware network by formulating an explicit contour loss function in the training process and achieved the best performance during the 2015 MICCAI Gland Segmentation (Glas) on-site challenge. Lu et al. [8] proposed BrcaSeg, a WSI processing pipeline that utilized deep learning to perform automatic segmentation and quantification of epithelial and stromal tissues for breast cancer WSI from TCGA. Besides the U-Net structure, Zhao [45] proposed a deep neural network, SCAU-Net, with spatial and channel attention for gland segmentation. SCAU-Net could effectively capture the nonlinear relationship between spatial-wise and channel-wise features, and achieve state-of-the-art gland segmentation performance. Moreover, with the help of the DeeplabV3 model, Musulin [44] developed an enhanced histopathology analysis tool that could accurately segment epithelial and stromal tissue for oral squamous cell carcinoma. Considering that the boundary of the gland is difficult to discriminate, Yan et al. [46] proposed a shape-aware adversarial deep learning framework, which had better tolerance to boundary uncertainty and was more effective for boundary detection. In addition, due to the fixed encoder-decoder structure, U-Net is not suitable for processing texture WSIs, Wen et al. [47] utilized a Gabor-based module to extract texture information at different scales and directions for tissue segmentation. Rojthoven et al. [48] proposed HookNet, a semantic segmentation model combining context information in WSIs via multiple branches of encoder-decoder CNN, for tissue segmentation.
Although much progress has been achieved, the superior performance of previous deep neural network-based methods mainly depends on the substantial number of training images with pixel-wise annotation, which are difficult to obtain due to the requirements of tremendous labeling efforts for experts. In order to reduce the overall labelling cost, several weakly supervised tissue segmentation algorithms have also been proposed [6][49][50]. For instance, Mahapatra [49] proposed a deep active learning framework that could actively select valuable samples from the unlabeled data for annotation, which significantly reduced the annotation efforts while still achieving comparable gland segmentation performance. Lai et al. [50] proposed a semi-supervised active learning framework with a region-based selection criterion. This framework iteratively selects regions for annotation queries to quickly expand the diversity and volume of the labeled set. Besides, Xie et al. [7] proposed a pairwise relation-based semi-supervised model for gland segmentation on histology images, which could produce considerable improvement in learning accuracy with limited labeled images and amounts of unlabeled images. Other studies include [6] having proposed a multiscale conditional GAN for epithelial region segmentation that could be used to compensate for the lack of labeled data in the training dataset. Moreover, Gupta et al. [51] introduced the idea of ‘image enrichment’ whereby the information content of images based on GAN is increased in order to enhance segmentation accuracy.
2. Cancer Diagnosis and Prognosis
Cancer is an aggressive disease with a low median survival rate. Ironically, the treatment process is long and very costly due to its high recurrence and mortality rates. Accurate early diagnosis and prognosis prediction of cancer is essential to enhance the patient’s survival rate [52][53]. It is now widely recognized that histopathological images are regarded as golden standards for the diagnosis and prognosis of human cancers [54][55]. Previous studies on histopathology image classification and prediction mainly focused on manual feature design. For instance, Cheng et al. [56] extracted a 150-dimensional handcrafted feature to describe each WSI, followed by the traditional classifiers to distinguish different types of renal cell carcinoma. Yu et al. [57] extracted 9879 quantitative features from each image tile and used regularized machine-learning methods to select the top features and to distinguish shorter-term survivors from longer-term survivors with adenocarcinoma or squamous cell carcinoma. Recently, with the success of deep learning in various computer vision tasks, training end-to-end deep learning models for various histopathological image analysis tasks without manually extracting features has drawn much attention [58][59][60].
Generally, the main challenge for applying deep learning algorithms for WSI classification and prediction is the large size of the WSI (e.g., 100,000 × 100,000 pixels), and it is impossible to directly feed these large images into the deep neural network for model training [61][62]. To address this challenge, there are two main lines of approaches, the patch-based and WSI-based methods (which are summarized in Figure 3).
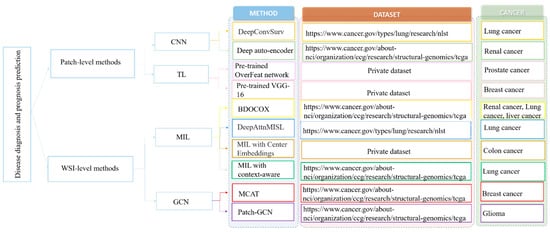
2.1. Patch-Level Methods
In connection with the large size of WSI, the patch-based methods required the pathologist to select the region of interests from WSI that are representative, then the selected regions were split into patches with a significantly smaller size for deep model training [63][64][72]. For instance, Zhu et al. [63] developed a deep CNN for survival analysis (DeepConvSurv) with the pathological patches derived from the WSI. They demonstrated that the end-to-end learning algorithm, DeepConvSurv, outperformed the standard Cox proportional hazard model. Cheng et al. [64] applied a deep autoencoder to aggregate the extracted patches into different groups and then learn topological features from the clusters to characterize cell distributions of different cell types for survival prediction.
By considering that training a model from scratch requires a very large dataset and takes a long time to train. Some patch-based methods also adopted the transfer learning model (TL) to speed up the training procedure, as well as improve the classification performance. TL provides an effective solution for feasibly and fast customized accurate models by transferring and fine-tuning the learned knowledge of pre-trained models over large datasets. For instance, Xu et al. [72] exploited CNN activation features to achieve region-level classification results. Specifically, they first over-segmented each preselected region into a set of overlapping patches. A TL strategy was then explored by pretraining CNN with ImageNet. Finally, an SVM classifier was adopted for classification. Similarly, Källénet et al. [65] extracted features from the divided patches via the pre-trained OverFeat network. The RF classifier was applied to discriminate the subtypes in prostatic adenocarcinoma. Moreover, in [66], the pre-trained VGG-16 network was first applied to extract descriptors from the preselected patches. Then, the feature representation of WSI was computed by the average pooling of the feature representations of its associated patches.
2.2. WSI-Level Methods
Although much progress has been achieved, the abovementioned patch-level prediction methods still have several inherent drawbacks. First, the patch-based methods required labor-sensitive patch-level annotation, which would increase the workload for the pathologist [73]. Second, most of the existing patch-based methods usually assumed that the diagnosis or survival information with each randomly selected patch was the same as its corresponding WSI, which neglected the fact that WSI usually had large heterogenous patterns and thus the patch-level label would not always match the WSI-level label [74].
In view of these challenges, building diagnosis/prognosis models only relying on WSI-level annotation has been widely investigated [67][74][75]. Among the WSI-based methods, the multi-instance learning (MIL) framework was a simple but most effective tool. For example, Shao et al. [67] considered the ordinal characteristic of the survival process by adding a ranking-based regularization term on the Cox model and used the average pooling strategies to aggregate the instance-level results to the WSI-level prediction results (Figure 4). Similarly, Iizuka et al. [75] first trained a CNN model using millions of tiles extracted from the WSI. Then, a max-pooling strategy combined with the recurrent neural network was adopted to fuse the patch-level results into WSI-level prediction results. However, by considering the simple decision fusion approaches (e.g., average pooling and max pooling) were insufficiently robust to make the right WSI-level prediction, Yao et al. [68] proposed an attention-guided deep multiple instance learning network (DeepAttnMISL) for survival prediction from WSI. In comparison with the traditional pooling strategies, attention-based aggregation is more flexible and adaptive for survival prediction. In addition, Chikontwe et al. [69] presented a novel MIL framework for histopathology slide classification. The proposed framework could be applied for both instance and bag level learning with a center loss that minimized intraclass distances in the embedding space. The experimental results also suggested that the proposed method could achieve overall improved performance over recent state-of-the-art methods. Moreover, Wang et al. [74] first extracted the spatial contextual features from each patch. Then, a globally holistic region descriptor was calculated after aggregating the features from multiple representative instances for WSI-level classification.
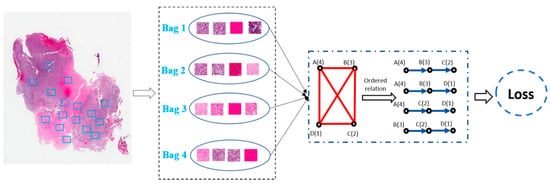
Figure 4. Weakly-supervised deep ordinal Cox model (BDOCOX) for survival prediction from WSI.
Although CNN-based MIL frameworks have shown impressive performance in the field of histopathology analysis, they are unable to capture complex neighborhood information as they analyze local areas determined by the convolutional kernel to extract interaction information between objects. Recently, some researchers have also applied the graph convolutional network (GCN) to analyze histopathological images for the diagnosis and prognosis of human cancers [70][76], which are becoming increasingly useful for medical diagnosis and prognosis. For instance, Chen et al. [70] presented a context-aware graph convolutional network that hierarchically aggregates instance-level histology features to model local- and global-level topological structures in the tumor microenvironment. Li et al. [76] proposed to model WSI as a graph and then develop a graph convolutional neural network with attention learning that better serves the survival prediction by rendering the optimal graph representations of WSIs. Moreover, the study in [77] presented a patch relevance-enhanced graph convolutional network (RGCN) to explicitly model the correlations of different patches in WSI, which can approximately estimate the diagnosis-related regions in WSI. Extensive experiments on real lung and brain carcinoma WSIs have demonstrated their effectiveness since GCNs can better exploit and preserve neighboring relations compared with CNN-based models. Besides, some researchers have noticed the relation between genes and images. Chen et al. [71] presented a multimodal co-attention transformer (MCAT) framework that learns an interpretable, dense co-attention mapping between WSI and genomic features formulated in an embedding space.
References
- Chan, L.; Hosseini, M.S.; Rowsell, C.; Plataniotis, K.N.; Damaskinos, S. Histosegnet: Semantic segmentation of histological tissue type in whole slide images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Thessaloniki, Greece, 23–25 September 2019; pp. 10662–10671.
- Zhang, H.; Liu, J.; Yu, Z.; Wang, P. MASG-GAN: A multi-view attention superpixel-guided generative adversarial network for efficient and simultaneous histopathology image segmentation and classification. Neurocomputing 2021, 463, 275–291.
- Sucher, R.; Sucher, E. Artificial intelligence is poised to revolutionize human liver allocation and decrease medical costs associated with liver transplantation. HepatoBiliary Surg. Nutr. 2020, 9, 679.
- Chen, H.; Qi, X.; Yu, L.; Dou, Q.; Qin, J.; Heng, P.A. DCAN: Deep contour-aware networks for object instance segmentation from histology images. Med. Image Anal. 2017, 36, 135–146.
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29.
- Li, W.; Li, J.; Polson, J.; Wang, Z.; Speier, W.; Arnold, C. High resolution histopathology image generation and segmentation through adversarial training. Med. Image Anal 2022, 75, 102251.
- Xie, Y.; Zhang, J.; Liao, Z.; Verjans, J.; Shen, C.; Xia, Y. Pairwise relation learning for semi-supervised gland segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Online, 4–8 October 2020; pp. 417–427.
- Lu, Z.; Zhan, X.; Wu, Y.; Cheng, J.; Shao, W.; Ni, D.; Han, Z.; Zhang, J.; Feng, Q.; Huang, K. BrcaSeg: A Deep Learning Approach for Tissue Quantification and Genomic Correlations of Histopathological Images. Genom. Proteom. Bioinform. 2021.
- Raza, S.E.A.; Cheung, L.; Epstein, D.; Pelengaris, S.; Khan, M.; Rajpoot, N.M. Mimo-net: A multi-input multi-output convolutional neural network for cell segmentation in fluorescence microscopy images. In Proceedings of the 2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017; pp. 337–340.
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018, 23, 181–193.e7.
- Samanta, P.; Raipuria, G.; Singhal, N. Context Aggregation Network For Semantic Labeling In Histopathology Images. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 673–676.
- Mahbod, A.; Schaefer, G.; Ellinger, I.; Ecker, R.; Smedby, Ö.; Wang, C. A two-stage U-Net algorithm for segmentation of nuclei in H&E-stained tissues. In Proceedings of the European Congress on Digital Pathology, Warwick, UK, 10–13 April 2019; pp. 75–82.
- Yang, L.; Ghosh, R.P.; Franklin, J.M.; Chen, S.; You, C.; Narayan, R.R.; Melcher, M.L.; Liphardt, J.T. NuSeT: A deep learning tool for reliably separating and analyzing crowded cells. PLoS Comput. Biol. 2020, 16, e1008193.
- Zhao, B.; Chen, X.; Li, Z.; Yu, Z.; Yao, S.; Yan, L.; Wang, Y.; Liu, Z.; Liang, C.; Han, C. Triple U-net: Hematoxylin-aware nuclei segmentation with progressive dense feature aggregation. Med. Image Anal. 2020, 65, 101786.
- Song, Y.; Zhang, L.; Chen, S.; Ni, D.; Lei, B.; Wang, T. Accurate Segmentation of Cervical Cytoplasm and Nuclei Based on Multiscale Convolutional Network and Graph Partitioning. IEEE Trans. Biomed. Eng. 2015, 62, 2421–2433.
- Zhou, Y.; Chang, H.; Barner, K.E.; Parvin, B. Nuclei segmentation via sparsity constrained convolutional regression. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; pp. 1284–1287.
- Ciresan, D.; Giusti, A.; Gambardella, L.; Schmidhuber, J. Deep neural networks segment neuronal membranes in electron microscopy images. Adv. Neural Inf Process. Syst. 2012, 25, 2843–2851.
- Qi, X.; Xing, F.; Foran, D.J.; Yang, L. Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans. Biomed. Eng. 2012, 59, 754–765.
- Veta, M.; van Diest, P.J.; Willems, S.M.; Wang, H.; Madabhushi, A.; Cruz-Roa, A.; Gonzalez, F.; Larsen, A.B.; Vestergaard, J.S.; Dahl, A.B.; et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med. Image Anal. 2015, 20, 237–248.
- Oren, A.; Fernandes, J. The Bethesda system for the reporting of cervical/vaginal cytology. J. Am. Osteopath. Assoc. 1991, 91, 476–479.
- Anoraganingrum, D. Cell segmentation with median filter and mathematical morphology operation. In Proceedings of the 10th International Conference on Image Analysis and Processing, Venice, Italy, 27–29 September 1999; pp. 1043–1046.
- Liu, T.; Li, G.; Nie, J.; Tarokh, A.; Zhou, X.; Guo, L.; Malicki, J.; Xia, W.; Wong, S.T. An automated method for cell detection in zebrafish. Neuroinformatics 2008, 6, 5–21.
- Lu, Z.; Carneiro, G.; Bradley, A.P. An improved joint optimization of multiple level set functions for the segmentation of overlapping cervical cells. IEEE Trans. Image Process. 2015, 24, 1261–1272.
- Dorini, L.B.; Minetto, R.; Leite, N.J. Semiautomatic white blood cell segmentation based on multiscale analysis. IEEE J. Biomed. Health Inform. 2012, 17, 250–256.
- Zhang, C.; Yarkony, J.; Hamprecht, F.A. Cell detection and segmentation using correlation clustering. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Boston, MA, USA, 14–18 September 2014; pp. 9–16.
- Bergeest, J.-P.; Rohr, K. Efficient globally optimal segmentation of cells in fluorescence microscopy images using level sets and convex energy functionals. Med. Image Anal. 2012, 16, 1436–1444.
- Sahara, K.; Paredes, A.Z.; Tsilimigras, D.I.; Sasaki, K.; Moro, A.; Hyer, J.M.; Mehta, R.; Farooq, S.A.; Wu, L.; Endo, I. Machine learning predicts unpredicted deaths with high accuracy following hepatopancreatic surgery. Hepatobiliary Surg. Nutr. 2021, 10, 20.
- Mahmood, F.; Borders, D.; Chen, R.J.; McKay, G.N.; Salimian, K.J.; Baras, A.; Durr, N.J. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans. Med. Imaging 2019, 39, 3257–3267.
- Liu, D.; Zhang, D.; Song, Y.; Zhang, C.; Zhang, F.; O’Donnell, L.; Cai, W. Nuclei Segmentation via a Deep Panoptic Model with Semantic Feature Fusion. In Proceedings of the 2019 International Joint Conference on Artificial Intelligence, Macao, China, 10–16 August 2019; pp. 861–868.
- Moris, D.; Shaw, B.I.; Ong, C.; Connor, A.; Samoylova, M.L.; Kesseli, S.J.; Abraham, N.; Gloria, J.; Schmitz, R.; Fitch, Z.W.; et al. A simple scoring system to estimate perioperative mortality following liver resection for primary liver malignancy—the Hepatectomy Risk Score (HeRS). Hepatobiliary Surg. Nutr. 2021, 10, 315–324.
- Schmitz, R.; Madesta, F.; Nielsen, M.; Krause, J.; Steurer, S.; Werner, R.; Rösch, T. Multi-scale fully convolutional neural networks for histopathology image segmentation: From nuclear aberrations to the global tissue architecture. Med. Image Anal. 2021, 70, 101996.
- Xing, F.; Xie, Y.; Yang, L. An automatic learning-based framework for robust nucleus segmentation. IEEE Trans. Med. Imaging 2015, 35, 550–566.
- Settouti, N.; Bechar, M.E.; Daho, M.E.; Chikh, M.A. An optimised pixel-based classification approach for automatic white blood cells segmentation. Int. J. Biomed. Eng. Technol. 2020, 32, 144–160.
- Sahasrabudhe, M.; Christodoulidis, S.; Salgado, R.; Michiels, S.; Loi, S.; André, F.; Paragios, N.; Vakalopoulou, M. Self-supervised nuclei segmentation in histopathological images using attention. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Online, 4–8 October 2020; pp. 393–402.
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440.
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241.
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Muller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010, 28, 105–113.
- Sirinukunwattana, K.; Pluim, J.P.W.; Chen, H.; Qi, X.; Heng, P.A.; Guo, Y.B.; Wang, L.Y.; Matuszewski, B.J.; Bruni, E.; Sanchez, U.; et al. Gland segmentation in colon histology images: The glas challenge contest. Med. Image Anal. 2017, 35, 489–502.
- Vukicevic, A.M.; Radovic, M.; Zabotti, A.; Milic, V.; Hocevar, A.; Callegher, S.Z.; De Lucia, O.; De Vita, S.; Filipovic, N. Deep learning segmentation of Primary Sjögren’s syndrome affected salivary glands from ultrasonography images. Comput. Biol. Med. 2021, 129, 104154.
- Gunduz-Demir, C.; Kandemir, M.; Tosun, A.B.; Sokmensuer, C. Automatic segmentation of colon glands using object-graphs. Med. Image Anal. 2010, 14, 1–12.
- Salvi, M.; Bosco, M.; Molinaro, L.; Gambella, A.; Papotti, M.; Acharya, U.R.; Molinari, F. A hybrid deep learning approach for gland segmentation in prostate histopathological images. Artif. Intell. Med. 2021, 115, 102076.
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012, 3, 153–173.
- Chen, H.; Qi, X.; Yu, L.; Heng, P.-A. DCAN: Deep contour-aware networks for accurate gland segmentation. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, Nevada, NV, USA, 26 June–1 July 2016; pp. 2487–2496.
- Musulin, J.; Stifanic, D.; Zulijani, A.; Cabov, T.; Dekanic, A.; Car, Z. An Enhanced Histopathology Analysis: An AI-Based System for Multiclass Grading of Oral Squamous Cell Carcinoma and Segmenting of Epithelial and Stromal Tissue. Cancers 2021, 13, 1784.
- Zhao, P.; Zhang, J.; Fang, W.; Deng, S. SCAU-Net: Spatial-Channel Attention U-Net for Gland Segmentation. Front. Bioeng. Biotechnol. 2020, 8, 670.
- Yan, Z.; Yang, X.; Cheng, K.T. Enabling a Single Deep Learning Model for Accurate Gland Instance Segmentation: A Shape-Aware Adversarial Learning Framework. IEEE Trans. Med. Imaging 2020, 39, 2176–2189.
- Wen, Z.; Feng, R.; Liu, J.; Li, Y.; Ying, S. GCSBA-Net: Gabor-Based and Cascade Squeeze Bi-Attention Network for Gland Segmentation. IEEE J. Biomed. Health Inform. 2020, 25, 1185–1196.
- van Rijthoven, M.; Balkenhol, M.; Siliņa, K.; van der Laak, J.; Ciompi, F. HookNet: Multi-resolution convolutional neural networks for semantic segmentation in histopathology whole-slide images. Med. Image Anal. 2021, 68, 101890.
- Mahapatra, D.; Poellinger, A.; Shao, L.; Reyes, M. Interpretability-Driven Sample Selection Using Self Supervised Learning for Disease Classification and Segmentation. IEEE Trans. Med. Imaging 2021, 40, 2548–2562.
- Lai, Z.; Wang, C.; Oliveira, L.C.; Dugger, B.N.; Cheung, S.-C.; Chuah, C.-N. Joint Semi-supervised and Active Learning for Segmentation of Gigapixel Pathology Images with Cost-Effective Labeling. In Proceedings of the Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 591–600.
- Gupta, L.; Klinkhammer, B.M.; Boor, P.; Merhof, D.; Gadermayr, M. GAN-based image enrichment in digital pathology boosts segmentation accuracy. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 631–639.
- Hu, A.; Razmjooy, N. Brain tumor diagnosis based on metaheuristics and deep learning. Int. J. Imaging Syst. Technol. 2021, 31, 657–669.
- Shen, X.; Zhao, H.; Jin, X.; Chen, J.; Yu, Z.; Ramen, K.; Zheng, X.; Wu, X.; Shan, Y.; Bai, J. Development and validation of a machine learning-based nomogram for prediction of intrahepatic cholangiocarcinoma in patients with intrahepatic lithiasis. Hepatobiliary Surg. Nutr. 2021, 10, 749.
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020, 471, 61–71.
- Lau, W.Y.; Wang, K.; Zhang, X.P.; Li, L.Q.; Wen, T.F.; Chen, M.S.; Jia, W.D.; Xu, L.; Shi, J.; Guo, W.X.; et al. A new staging system for hepatocellular carcinoma associated with portal vein tumor thrombus. Hepatobiliary Surg. Nutr. 2021, 10, 782–795.
- Cheng, J.; Han, Z.; Mehra, R.; Shao, W.; Cheng, M.; Feng, Q.; Ni, D.; Huang, K.; Cheng, L.; Zhang, J. Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11. 2 translocation renal cell carcinoma. Nat. Commun. 2020, 11, 1778.
- Yu, K.H.; Zhang, C.; Berry, G.J.; Altman, R.B.; Re, C.; Rubin, D.L.; Snyder, M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016, 7, 12474.
- Hosseini, M.S.; Brawley-Hayes, J.A.; Zhang, Y.; Chan, L.; Plataniotis, K.N.; Damaskinos, S. Focus quality assessment of high-throughput whole slide imaging in digital pathology. IEEE Trans. Med. Imaging 2019, 39, 62–74.
- Yao, J.; Zhu, X.; Huang, J. Deep multi-instance learning for survival prediction from whole slide images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 496–504.
- Yang, H.; Kim, J.-Y.; Kim, H.; Adhikari, S.P. Guided soft attention network for classification of breast cancer histopathology images. IEEE Trans. Med. Imaging 2019, 39, 1306–1315.
- Chen, C.; Lu, M.Y.; Williamson, D.F.; Chen, T.Y.; Schaumberg, A.J.; Mahmood, F. Fast and Scalable Image Search For Histology. arXiv 2021, arXiv:2107.13587.
- Sun, H.; Zeng, X.; Xu, T.; Peng, G.; Ma, Y. Computer-aided diagnosis in histopathological images of the endometrium using a convolutional neural network and attention mechanisms. IEEE J. Biomed. Health Inform. 2019, 24, 1664–1676.
- Zhu, X.; Yao, J.; Huang, J. Deep convolutional neural network for survival analysis with pathological images. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016; pp. 544–547.
- Cheng, J.; Mo, X.; Wang, X.; Parwani, A.; Feng, Q.; Huang, K. Identification of topological features in renal tumor microenvironment associated with patient survival. Bioinformatics 2018, 34, 1024–1030.
- Källén, H.; Molin, J.; Heyden, A.; Lundström, C.; Åström, K. Towards grading gleason score using generically trained deep convolutional neural networks. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1163–1167.
- Mercan, C.; Aksoy, S.; Mercan, E.; Shapiro, L.G.; Weaver, D.L.; Elmore, J.G. From patch-level to ROI-level deep feature representations for breast histopathology classification. In Proceedings of the Medical Imaging 2019: Digital Pathology, San Diego, CA, USA, 20–21 February 2019; p. 109560H.
- Shao, W.; Wang, T.; Huang, Z.; Han, Z.; Zhang, J.; Huang, K. Weakly supervised deep ordinal cox model for survival prediction from whole-slide pathological images. IEEE Trans. Med. Imaging 2021, 40, 3739–3747.
- Yao, J.; Zhu, X.; Jonnagaddala, J.; Hawkins, N.; Huang, J. Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Med. Image Anal. 2020, 65, 101789.
- Chikontwe, P.; Kim, M.; Nam, S.J.; Go, H.; Park, S.H. Multiple instance learning with center embeddings for histopathology classification. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 519–528.
- Chen, R.J.; Lu, M.Y.; Shaban, M.; Chen, C.; Chen, T.Y.; Williamson, D.F.; Mahmood, F. Whole Slide Images are 2D Point Clouds: Context-Aware Survival Prediction using Patch-based Graph Convolutional Networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; pp. 339–349.
- Chen, R.J.; Lu, M.Y.; Weng, W.-H.; Chen, T.Y.; Williamson, D.F.; Manz, T.; Shady, M.; Mahmood, F. Multimodal Co-Attention Transformer for Survival Prediction in Gigapixel Whole Slide Images. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 11–17 October 2021; pp. 4015–4025.
- Xu, Y.; Jia, Z.; Wang, L.B.; Ai, Y.; Zhang, F.; Lai, M.; Chang, E.I. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 281.
- Marini, N.; Otalora, S.; Muller, H.; Atzori, M. Semi-supervised training of deep convolutional neural networks with heterogeneous data and few local annotations: An experiment on prostate histopathology image classification. Med. Image Anal. 2021, 73, 102165.
- Wang, X.; Chen, H.; Gan, C.; Lin, H.; Dou, Q.; Tsougenis, E.; Huang, Q.; Cai, M.; Heng, P.A. Weakly Supervised Deep Learning for Whole Slide Lung Cancer Image Analysis. IEEE Trans. Cybern. 2020, 50, 3950–3962.
- Iizuka, O.; Kanavati, F.; Kato, K.; Rambeau, M.; Arihiro, K.; Tsuneki, M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. Sci. Rep. 2020, 10, 1504.
- Li, R.; Yao, J.; Zhu, X.; Li, Y.; Huang, J. Graph CNN for survival analysis on whole slide pathological images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 174–182.
- Chen, Z.; Zhang, J.; Che, S.; Huang, J.; Han, X.; Yuan, Y. Diagnose Like A Pathologist: Weakly-Supervised Pathologist-Tree Network for Slide-Level Immunohistochemical Scoring. In Proceedings of the 35th AAAI Conference on Artificial Intelligence (AAAI-21), Online, 2–9 February 2021; pp. 47–54.
More
