Asthma is a respiratory condition often stemming from childhood, characterized by difficulty breathing and/or chest tightness. Current treatment options for both adults and children include beta-2 agonists, inhaled corticosteroids (ICS), and leukotriene modifiers (LTM). Despite recommendations by the Global Initiative for Asthma, a substantial number of patients are unresponsive to treatment and unable to control symptoms. Pharmacogenomics have increasingly become the front line of precision medicine, especially with the recent use of candidate gene and genome- wide association studies (GWAS). Screening patients preemptively could likely decrease adverse events and therapeutic failure. However, research in asthma, specifically in pediatrics, has been low. Although numerous adult trials have evaluated the impact of pharmacogenomics and treatment response, the lack of evidence in children has hindered progress towards clinical application.
- pediatric asthma
- pharmacogenomics
- beta-2 receptors
- inhaled corticosteroids
- leukotriene modifiers
1. Introduction
2. Inhaled Corticosteroids (ICS)
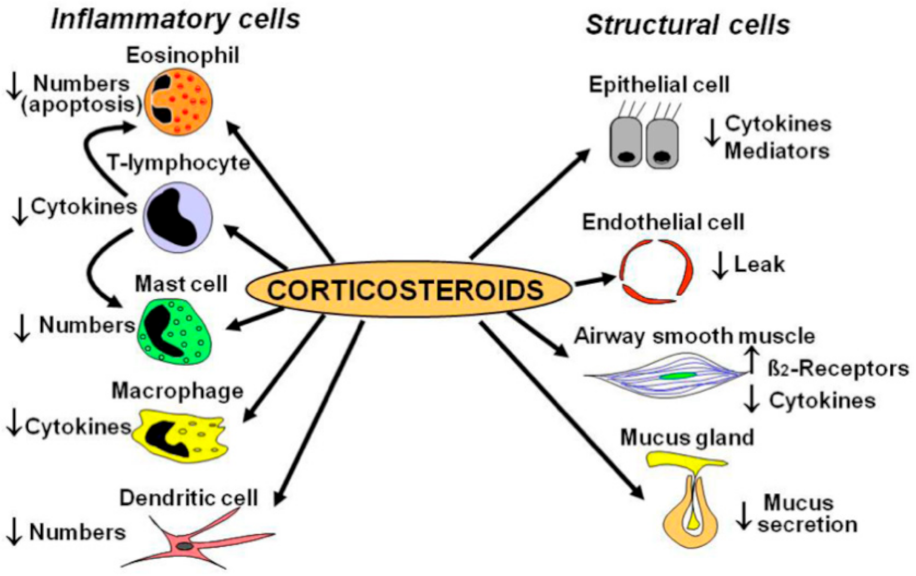 Figure 1. ICS Mechanism of Action. Adapted from [11].
Figure 1. ICS Mechanism of Action. Adapted from [11].| Study | Population | Gene | Medication | Measurement | Results |
|---|---|---|---|---|---|
| Tantisira [12] | White | GLCCI1 | Budesonide | Change in FEV1 from baseline | |
| CYP3A4 *22 produced favorable response | |||||
| Stockman | |||||
| [ | |||||
| 23 | |||||
| ] | |||||
| White | |||||
| CYP3A4, CYP3A5, and CYP3A7 | Beclomethasone | Asthma control scores | CYP3A5 *3/*3 produced favorable response |
3. Leukotriene Modifiers (LTM)
3.1. Genes That Affect Montelukast Response
| Study | Population | Gene | Measurement | Results |
|---|
| rs37972 TT and rs37973 G allele variants showed unfavorable responses |
| Kim [28] | Korean | TBXA2 | FEV1 | Combination of +795 CT/CC and +924 TT showed unfavorable response | ||||
| Thompson [13] | European | GLCCI1 | ICS | Exacerbation (hospital visits and oral steroid use) | rs37973 G allele showed unfavorable response | |||
| Vijverberg [14] | Europe | GLCCI1 | Budesonide | Exacerbation (emergency room visits, hospital visits, oral steroid use) | No association found at rs37972 | |||
| Huang [15] | Chinese | GLCCI1 | ICS | MMEF | rs37969, rs37972, and rs37973 produced favorable response. | |||
| Kim [16] | Korean | HDAC1 | ||||||
| Klotsman [29] | Multiethnic | ALOX5 CystLTR2 LTC4S |
PEF PEF FEV1 and PEF |
rs4987105 TT and rs4986832 AA variants showed favorable outcomes rs912277 TT and rs912278 TC variants produced favorable response No association found at rs730012 |
||||
| Telleria [30] | Spain | ALOX5 | % change in FEV1, exacerbations, and rescue inhaler need | rs59439148 5/5 and 5/4 copies showed favorable outcomes | ||||
| Kang [31] | Korean | LTC4S PTGDR |
≥10% increase in FEV1 post exercise challenge | No significance at rs730012 rs803010 TT produced favorable response |
ICS | % change in FEV1 pre- and post-bronchodilator | rs1741981 CT and TT produced favorable response | |
| Lee [32] | Korean | LTC4S CystLTR1 |
>10% increase in FEV1 post exercise challenge | No significance at SNP rs730012 No significance found |
Tantisira [17] | |||
| Whelan [33] | Caucasians | CRHR1 | Budesonide | % change in FEV1 from baseline | African American and Caucasianrs242941T produced favorable response | |||
| LTC4S | FENO | rs730012 AC produced favorable response | Tantisira [18] | Multiethnic | TBX21 | Budesonide | % change in FEV1 and PC20 | rs2240017Q produced favorable response |
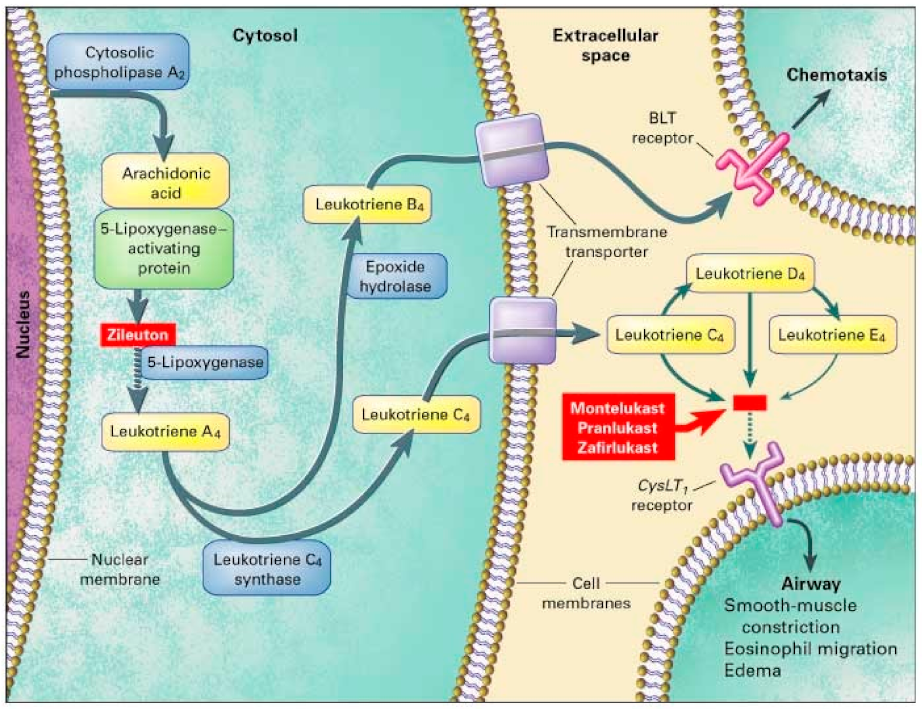 Figure 2. Leukotriene Modifiers Mechanism of Action. Adapted from [36].
Figure 2. Leukotriene Modifiers Mechanism of Action. Adapted from [36].| Study | Population | Gene | Medication | Measurement | Results | ||||
|---|---|---|---|---|---|---|---|---|---|
| Lipworth [37] | Scotland | ADRB2 | Fluticasone + montelukast or salmeterol + fluticasone | School absences, FEV1, asthma symptoms | Arg16Arg produced favorable responses to fluticasone and montelukast | ||||
| Mougey | |||||||||
| [ | |||||||||
| 34 | |||||||||
| ] | |||||||||
| Multiethnic | SLCO2B1 | Asthma symptom utility index (ASUI) | rs12422149 GG produced favorable response | No association at rs2306168 |
|||||
| Tantisira [19] | Multiethnic | FCER2 | Budesonide | Exacerbations (ER visits or hospitalization) | rs28364072 CC produced unfavorable response | ||||
| Li [35] | Chinese | SLCO2B1CYP2C8 | Drug clearance | Koster [20] | European | FCER2 | ICS, ICS + salmeterol, ICS + salmeterol + montelukast | Exacerbations (hospital visits and/or oral steroid use) | rs28364072 CC produced unfavorable response |
| Decreased clearance in rs12422149 GG No association found | Keskin [21] | Turkey | NR3C1 | Fluticasone | FEV1 improvement at 4 h | rs41423247 GG produced favorable response | |||
| Stockman [22] | Whites | CYP3A4, CYP3A5, and CYP3A7 | Fluticasone | Asthma control scores |
3.2. Genes Affecting Other Leukotriene Modifier Responses
| Study | Population | Gene | Medication | Measurement | Results |
|---|---|---|---|---|---|
| Tcheurekdjlan [41] | Mexican and Puerto Ricans | LTA4H ALOX5AP |
Albuterol + leukotriene modifiers (montelukast, zafirlukast, and zileuton) | % change in FEV1 pre and post bronchodilator | LTA4H rs2540491 A variants and rs2540487 GA variants produced favorable response No association of ALOX5AP variants alone rs10507391 A allele and rs9551963 C allele magnifies LTA4H response |
References
- Centers for Disease Control and Prevention. 2019 National Health Interview Survey (NHIS) Data. Available online: https://www.cdc.gov/asthma/nhis/2019/data.htm/ (accessed on 25 October 2021).
- Weiss, S.T.; Litonjua, A.A.; Lange, C.; Lazarus, R.; Liggett, S.B.; Bleecker, E.R.; Tantisira, K.G. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics 2006, 6, 311–326.
- The Global Asthma Report 2018; Global Asthma Network: Auckland, New Zealand. 2018. Available online: globalasthmareport.org (accessed on 31 January 2022).
- Nurmagambetov, T.; Kuwahara, R.; Garbe, P. The Economic Burden of Asthma in the United States 2008-2013. Ann. Am. Thorac Soc. 2018, 15, 348–356.
- Centers for Disease Control and Prevention. Asthma-Related Missed School Days among Children Aged 5–17 Years. Available online: https://www.cdc.gov/asthma/asthma_stats/missing_days.htm/ (accessed on 20 August 2021).
- Dharmage, S.C.; Perret, J.L.; Custovic, A. Epidemiology of Asthma in Children and Adults. Front Pediatr. 2019, 18, 246.
- Global Initiative for Asthma. Global Initiative for Asthma—GINA. Available online: https://ginasthma.org/ (accessed on 2 September 2021).
- Gustafsson, P.M.; Watson, L.; Davis, K.J.; Rabe, K.F. Poor asthma control in children: Evidence from epidemiological surveys and implications for clinical practice. Int. J. Clin. Pract. 2006, 60, 321–334.
- Gallagher, R.M.; Mason, J.R.; Bird, K.A.; Kirkham, J.J.; Peak, M.; Williamson, P.R.; Nunn, A.J.; Turner, M.A.; Pirmohamed, M.; Smyth, R.L. Adverse drug reactions causing admission to a paediatric hospital. PLoS ONE 2012, 7, e50127.
- Clancy, J.P.; Johnson, S.G.; Yee, S.W.; McDonagh, E.M.; Caudle, K.E.; Klein, T.E.; Cannavo, M.; Giacomini, K.M. Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for ivacaftor therapy in the context of CFTR genotype. Clin. Pharmacol. Ther. 2014, 95, 592–597.
- Barnes, P.J. Inhaled Corticosteroids. Pharmaceuticals 2010, 3, 514–540.
- Tantisira, K.G.; Lasky-Su, J.; Harada, M.; Murphy, A.; Litonjua, A.A.; Himes, B.E.; Lange, C.; Lazarus, R.; Sylvia, J.; Klanderman, B.; et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N. Engl. J. Med. 2011, 365, 1173–1183.
- Thompson, B.; Hawcutt, D.; Carr, D.; Jorgensen, A.; Smyth, R.; Pirmohamed, M. S31 variation at GLC1CI1: Association with increased steroid dose but not adrenal suppression in asthmatic children. Thorax 2012, 67 (Suppl. 2), A17.
- Vijverberg, S.J.; Tavendale, R.; Leusink, M.; Koenderman, L.; Raaijmakers, J.A.; Postma, D.S.; Koppelman, G.H.; Turner, S.W.; Mukhopadhyay, S.; Palmer, C.N.; et al. Pharmacogenetic analysis of GLCCI1 in three north European pediatric asthma populations with a reported use of inhaled corticosteroids. Pharmacogenomics 2014, 15, 799–806.
- Huang, J.; Hu, X.; Zheng, X.; Kuang, J.; Liu, C.; Wang, X.; Tang, Y. Effects of STIP1 and GLCCI1 polymorphisms on the risk of childhood asthma and inhaled corticosteroid response in Chinese asthmatic children. BMC Pulm. Med. 2020, 20, 303.
- Kim, M.H.; Kim, S.H.; Kim, Y.K.; Hong, S.J.; Min, K.U.; Cho, S.H.; Park, H.W. A polymorphism in the histone deacetylase 1 gene is associated with the response to corticosteroids in asthmatics. Korean J. Intern. Med. 2013, 28, 708–714.
- Tantisira, K.G.; Lake, S.; Silverman, E.S.; Palmer, L.J.; Lazarus, R.; Silverman, E.K.; Liggett, S.B.; Gelfand, E.W.; Rosenwasser, L.J.; Richter, B.; et al. Corticosteroid pharmacogenetics: Association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum. Mol. Genet. 2004, 13, 1353–1359.
- Tantisira, K.G.; Hwang, E.S.; Raby, B.A.; Silverman, E.S.; Lake, S.L.; Richter, B.G.; Peng, S.L.; Drazen, J.M.; Glimcher, L.H.; Weiss, S.T. TBX21: A functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc. Natl. Acad. Sci. USA 2004, 101, 18099–18104.
- Tantisira, K.G.; Silverman, E.S.; Mariani, T.J.; Xu, J.; Richter, B.G.; Klanderman, B.J.; Litonjua, A.A.; Lazarus, R.; Rosenwasser, L.J.; Fuhlbrigge, A.L.; et al. FCER2: A pharmacogenetic basis for severe exacerbations in children with asthma. J. Allergy. Clin. Immunol. 2007, 120, 1285–1291.
- Koster, E.S.; Maitland-van der Zee, A.H.; Tavendale, R.; Mukhopadhyay, S.; Vijverberg, S.J.; Raaijmakers, J.A.; Palmer, C.N. FCER2 T2206C variant associated with chronic symptoms and exacerbations in steroid-treated asthmatic children. Allergy 2011, 66, 1546–1552.
- Keskin, O.; Uluca, Ü.; Birben, E.; Coşkun, Y.; Ozkars, M.Y.; Keskin, M.; Kucukosmanoglu, E.; Kalayci, O. Genetic associations of the response to inhaled corticosteroids in children during an asthma exacerbation. Pediatr. Allergy Immunol. 2016, 27, 507–513.
- Stockmann, C.; Fassl, B.; Gaedigk, R.; Nkoy, F.; Uchida, D.A.; Monson, S.; Reilly, C.A.; Leeder, J.S.; Yost, G.S.; Ward, R.M. Fluticasone propionate pharmacogenetics: CYP3A4*22 polymorphism and pediatric asthma control. J. Pediatr. 2013, 162, 1227–1227, 1227.e1–2.
- Stockmann, C.; Reilly, C.A.; Fassl, B.; Gaedigk, R.; Nkoy, F.; Stone, B.; Roberts, J.K.; Uchida, D.A.; Leeder, J.S.; Sherwin, C.M.; et al. Effect of CYP3A5*3 on asthma control among children treated with inhaled beclomethasone. J. Allergy Clin. Immunol. 2015, 136, 505–507.
- Lima, J.J. Treatment heterogeneity in asthma: Genetics of response to leukotriene modifiers. Mol. Diagn. Ther. 2007, 11, 97–104.
- Choi, J. Leukotriene Receptor Antagonists. Available online: https://www.ncbi.nlm.nih.gov/books/NBK554445/ (accessed on 5 November 2021).
- Drazen, J.M.; Israel, E.; O’Byrne, P.M. Treatment of asthma with drugs modifying the leukotriene pathway. N. Engl. J. Med. 1999, 340, 197–206.
- Bäck, M. Functional characteristics of cysteinyl-leukotriene receptor subtypes. Life Sci. 2002, 71, 611–622.
- Kim, J.H.; Lee, S.Y.; Kim, H.B.; Jin, H.S.; Yu, J.H.; Kim, B.J.; Kim, B.S.; Kang, M.J.; Jang, S.O.; Hong, S.J. TBXA2R gene polymorphism and responsiveness to leukotriene receptor antagonist in children with asthma. Clin. Exp. Allergy 2008, 38, 51–59.
- Klotsman, M.; York, T.P.; Pillai, S.G.; Vargas-Irwin, C.; Sharma, S.S.; van den Oord, E.J.; Anderson, W.H. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet. Genom. 2007, 17, 189–196.
- Telleria, J.J.; Blanco-Quiros, A.; Varillas, D.; Armentia, A.; Fernandez-Carvajal, I.; Jesus Alonso, M.; Diez, I. ALOX5 promoter genotype and response to montelukast in moderate persistent asthma. Respir. Med. 2008, 102, 857–861.
- Kang, M.J.; Kwon, J.W.; Kim, B.J.; Yu, J.; Choi, W.A.; Shin, Y.J.; Hong, S.J. Polymorphisms of the PTGDR and LTC4S influence responsiveness to leukotriene receptor antagonists in Korean children with asthma. J. Hum. Genet. 2011, 56, 284–289.
- Lee, S.Y.; Kim, H.B.; Kim, J.H.; Kim, B.S.; Kang, M.J.; Jang, S.O.; Seo, H.J.; Hong, S.J. Responsiveness to montelukast is associated with bronchial hyperresponsiveness and total immunoglobulin E but not polymorphisms in the leukotriene C4 synthase and cysteinyl leukotriene receptor 1 genes in Korean children with exercise-induced asthma (EIA). Clin. Exp. Allergy 2007, 37, 1487–1493.
- Whelan, G.J.; Blake, K.; Kissoon, N.; Duckworth, L.J.; Wang, J.; Sylvester, J.E.; Lima, J.J. Effect of montelukast on time-course of exhaled nitric oxide in asthma: Influence of LTC4 synthase A(-444)C polymorphism. Pediatr Pulmonol. 2003, 36, 413–420.
- Mougey, E.B.; Feng, H.; Castro, M.; Irvin, C.G.; Lima, J.J. Absorption of montelukast is transporter mediated: A common variant of OATP2B1 is associated with reduced plasma concentrations and poor response. Pharmacogenet. Genom. 2009, 19, 129–138.
- Li, Q.; Wang, K.; Shi, H.Y.; Wu, Y.E.; Zhou, Y.; Kan, M.; Zheng, Y.; Hao, G.X.; Yang, X.M.; Yang, Y.L. Developmental Pharmacogenetics of SLCO2B1 on Montelukast Pharmacokinetics in Chinese Children. Drug Des. Devel. Ther. 2019, 13, 4405–4411.
- Mougey, E.; Lang, J.E.; Allayee, H.; Teague, W.G.; Dozor, A.J.; Wise, R.A.; Lima, J.J. ALOX5 polymorphism associates with increased leukotriene production and reduced lung function and asthma control in children with poorly controlled asthma. Clin. Exp. Allergy 2013, 43, 512–520.
- Lipworth, B.J.; Basu, K.; Donald, H.P.; Tavendale, R.; Macgregor, D.F.; Ogston, S.A.; Palmer, C.N.; Mukhopadhyay, S. Tailored second-line therapy in asthmatic children with the Arg(16) genotype. Clin. Sci. (Lond.). 2013, 124, 521–528.
- Karonen, T.; Neuvonen, P.J.; Backman, J.T. CYP2C8 but not CYP3A4 is important in the pharmacokinetics of montelukast. Br. J. Clin. Pharmacol. 2012, 73, 257–267.
- Aquilante, C.L.; Niemi, M.; Gong, L.; Altman, R.B.; Klein, T.E. PharmGKB summary: Very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 8. Pharmacogenet. Genom. 2013, 23, 721–728.
- Bouchette, D.; Preuss, C.V. Zileuton. Available online: https://www.ncbi.nlm.nih.gov/books/NBK448202/ (accessed on 21 August 2021).
- Tcheurekdjian, H.; Via, M.; De Giacomo, A.; Corvol, H.; Eng, C.; Thyne, S.; Chapela, R.; Rodriguez-Cintron, W.; Rodriguez-Santana, J.R.; Avila, P.C.; et al. Genetics of Asthma in Latino Americans Study. Genetics of asthma in Latino Americans study. ALOX5AP and LTA4H polymorphisms modify augmentation of bronchodilator responsiveness by leukotriene modifiers in Latinos. J. Allergy Clin. Immunol. 2010, 126, 853–858.
