Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Nora Tang and Version 1 by Ewa Ostrycharz.
The complement system (CS) is part of the human immune system, consisting of more than 30 proteins that play a vital role in the protection against various pathogens and diseases, including viral diseases. Activated via three pathways, the classical pathway (CP), the lectin pathway (LP), and the alternative pathway (AP), the complement system leads to the formation of a membrane attack complex (MAC) that disrupts the membrane of target cells, leading to cell lysis and death. Due to the increasing number of reports on its role in viral diseases, which may have implications for research on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
- complement system
- viral diseases
- respiratory diseases
1. The Complement System (CS)
The complement system (CS) plays a key role in the defense against pathogens as part of the human immune system [1,2][1][2]. It consists of more than 30 soluble proteins synthesized mainly by the liver (but also by leukocytes), which are present in the plasma in an inactive form [3], as well as membrane-bound regulators and receptors that interact with various cells and mediators of the immune system [4]. Activation of the complement system involves a cascade of enzymatic and non-enzymatic reactions that culminate in opsonization by various opsonins (e.g., C3b and C4b) of the pathogens or pathogen-virus-infected cells and then lysis of these cells by a set of proteins that form a membrane attack complex (MAC) [5,6][5][6]. In addition, activation of the complement system leads to the production of anaphylatoxins—potent proinflammatory molecules. Complement also served to mediate clearance of immune complexes and damaged self cells or cell debris and to mediate phagocytosis by neutrophils and monocytes [7,8][7][8]. A limited number of reports indicate that complement may contribute to the regulation of the anti-inflammatory response. This process involves T lymphocytes (Treg), crucial in the production of anti-inflammatory cytokines such as transforming growth factor-β (TGF-β), IL-10, and IL-35 [8]. The system is activated by three main pathways: the classical pathway (CP), the lectin pathway (LP), and the alternative pathway (AP) (Figure 1) [9].
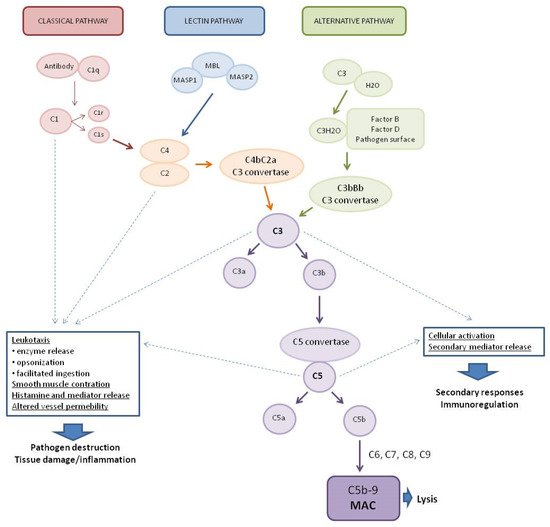
Figure 1. Pathways of complement system activation. MASP1—mannan-binding lectin associated serine protease-1, MASP2—mannan-binding lectin associated serine protease-2, MBL—mannose-binding lectin, and MAC—membrane attack complex.
The classical complement system pathway is referred to as “antibody-dependent” because of the involvement of IgM/IgG antibodies in the activation. Protein C1q (part of the C1 complex which consists of six molecules of C1q, two molecules of C1r, and two molecules of C1s) binds to the Fc region of complement-fixing antibodies (generally IgG1 and IgM) attached to pathogenic surfaces and pathogen-infected cells. This results in the activation of C1r and C1s proteases in the C1 complex [10,11][10][11]. C1s cleaves C4 and C2 proteases into large fragments (C4b, C2a) and small fragments (C4a, C2b). The larger fragments combine to form the C4bC2a complex on the pathogen surface which leads to the cleavage of C3 into anaphylatoxin C3a and opsonin C3b. The production of C3 convertase is the point where three complement pathways converge and afterwards have common steps to form MAC [1]. The lectin complement system pathway is similar to the classical pathway, but is independent of immunoglobulins. It does not recognize antigen antibody complexes but employs germline-encoded pattern-recognition receptors (PRRs) such as mannose-binding lectin (MBL) and ficolins [5]. When MBL recognizes and binds to carbohydrates in pathogen-associated molecular patterns (PAMPs), such as those found in viruses [12], MBL-associated serine proteases (MASPs, MBL-associated serine protease-2 (MASP-2), and MBL-associated serine protease-1 (MASP-1)) are activated to cleave complement components C2 and C4, which then leads to the generation of C3 convertase [13,14,15][13][14][15]. The alternative complement system pathway, in contrast to the classical and lectin pathways, consists of three processes that partially overlap. In the alternative pathway, activators include viruses [16]. Activation of the C3 convertase in this pathway occurs slowly in plasma and lead to the formation of C3H2O. C3 activation is induced by the presence of various surfaces which lack complement regulatory proteins which adsorb C3 to the surface to induce its conformational changes. C3H2O can then bind factor B (FB) to induce another conformational change as FB is cleaved into two components (FBb and FBa) by factor D (FD) [5]. This convertase begins to cleave C3 into C3a and C3b, in a manner analogous to the C4bC2a convertase in the classical and lectin pathway. The resulting C3b can bind to cell surfaces and FB to form the predominant convertase in the alternative pathway, i.e., C3bBb [2,17][2][17]. This C3bBb complex can be further stabilized by properdin, protecting it from the inactivating factor H (FH) and factor I (FI) [18]. All surface-bound C3 convertases, regardless of their origin, can induce an amplification branch of the alternative complement system pathway through the activation of C3 [19], which increases the density of deposited C3b and gradually leads to the formation of convertases that contain an additional C3b molecule (C4b2b3b or C3bBb3b) shifting its substrate specificity towards C5. The C5 convertase of the alternative pathway cleaves C5 into the anaphylatoxin C5a and the C5b fragment. When C5b binds to C6 and C7, the complex is inserted into cell membranes and interacts with C8, inducing the binding of several C9 units to form the MAC complex C5b6789 [20].
Activation of the complement system, regardless of the pathway, results in the generation of three broad effector pathways that enable the complement system to perform its physiological functions in host defense: direct lysis of target surfaces by the MAC, alerting and stimulating the immune system by producing potent proinflammatory anaphylatoxins, and opsonization of target surfaces by opsonins C4b, C3b, and C3bi [2,5,17][2][5][17]. MAC formation and targeted lysis are important effectors of the complement system’s anti-pathogenic actions [9]. Some cleavage products and the complement system activation products can act as anaphylatoxins and have broader immune regulatory functions. Most notably, the cleavage products C3a and C5a can be generated by all three pathways and can act as potent immune regulators, whereas C4a is generated exclusively by the classical and lectin pathways [21]. C3a and C5a can affect chemotaxis of eosinophilia, fibroblasts, macrophages, mast cells, and monocytes to the site of infection and inflammation, C5a alone is responsible for neutrophil recruitment [6], while C4a acts as an effector protein and increases endothelial cell permeability and enhances stress fiber formation [22]. In addition to their roles in chemotaxis, C3a and C5a have been implicated in the regulation of vasodilation, increased vascular permeability [23], and the production of various cytokines, including IL-1β, IL-8/CXCL-8, CCL5, IL-6, and tumor necrosis factor-α (TNF-α) [24,25][24][25].
The complement system, with its ability to form channels in the cell membrane, induce phagocytosis, and cause mast cell degranulation, is a dangerous weapon, and without constant supervision by the various mechanisms regulating its activity (Table 1), it could easily lead to cell and tissue damage in our body.
Table 1. Factors regulating the activity of the complement.
Factors regulating the activity of the complement.
| Factor | Function | References | ||||||
|---|---|---|---|---|---|---|---|---|
| C1 Esterase Inhibitor (C1-INH) | Plasma serine proteinase inhibitor (serpin). Binds to activated C1r and C1s, irreversibly inhibiting their activity, inhibition of classical pathway. Inhibits MASP-1 and MASP-2 | [26][27] | ||||||
| Factor I (FI) | Protease inactivating C4b and C3b with cofactors | [28] | ||||||
| Soluble Regulatory Proteins: C4b-Binding Protein and factor H (FH) | Cofactors for factor I. Accelerates the decomposition of the C4b2a and C3bBb complex. It is necessary for the regulation of C3 activity | [27][29][30][31] | ||||||
| Membrane Regulatory Proteins | Protect cells from complement mediated lysis | [28][32] | ||||||
| Decay-accelerating Factor (DAF) (CD55) | Factor accelerating the decomposition of C3 and C5 convertases | |||||||
| Membrane cofactor protein (MCP, CD46) |
Binds components C3b and C4b in the free state or in convertase. | |||||||
| Flaviviridae | Non-structural protein NS1 function as a regulator of the complement system. NS1 directly binds C4b binding protein (C4BP) on the surface of infected cells resulting in inhibition of complement activation in all pathways and MAC formation | [66] | Properdin | Stabilizes C3 and C5 convertases | [33][ | |||
| Dengue | 34] | |||||||
| Dengue virus (DENV 1-4) | NS1 competitively binds to MBL, which prevents the later from recognizing and neutralizing the virus. NS1 binds clusterin/vitronectin on the surface of infected cells, resulting in the inhibition of complement activation in all pathways and MAC formation |
[67][68][69] | Soluble MAC Inhibitors | [27][32] | ||||
| Zika Virus Disease (ZVD) | Vitronectin | Binds MAC and prevents the complex from being inserted into the cell membrane | ||||||
| Clusterin | Inactivates MAC with vitronectin | |||||||
| Membrane MAC Inhibitor CD59 | The primary membrane-bound inhibitor of the MAC. It binds to C8 and C9, preventing the incorporation and polymerization of C9 | [27][32] | ||||||
2. The Complement System at the Crossroads of the Innate and Adaptive Immune Response
The complement system is a part of the immune system and plays the role of a functional bridge between innate and adaptive immune responses that allows an integrated host defense against pathogens [9]. The humoral immune response is designed to protect extracellular spaces by activating effector and memory B cells and producing antibodies, leading to the neutralization and opsonization of the pathogen and providing immune memory against reinfection [5]. Complement system effectors are involved in humoral responses at multiple stages of B lymphocyte differentiation [35]. The complement system enhances B cell immunity mainly through complement receptors (CR), complement receptor type 1 (CR1) (CD35), and complement receptor type 1 (CR2) (CD21) expressed on B lymphocytes and follicular dendritic cells (FDCs). The CR2 receptor forms a receptor complex with the signaling protein CD19 and protein CD81 to form a receptor complex (CD21-CD19-CD81) of B lymphocytes. This supports enhanced B cell receptor (BCR e.g., surface immunoglobulins)-mediated signaling upon encountering a pathogen coated with complement system opsonins, resulting in a lowered threshold for B lymphocyte activation [36,37][36][37]. Coupling C3d to a low-affinity antigen which in the absence of coupling would cause B cell death, results not only in survival but also in B cell activation and antibody production [38]. Furthermore, the CR2 receptor mediates antigen-independent signals that are essential for B cell survival [39].
A regulatory effect on B lymphocytes is also induced by anaphylatoxins. Anaphylatoxin C3a causes the suppression of polyclonal B lymphocyte responses, while C5a promotes naive B cell migration and memory [40,41][40][41]. The complement system also influences T cell-associated responses. Experiments on mice lacking complement system inhibitory proteins (Decay-accelerating Factor (DAF) and CD59) highlight the important regulatory role of the complement system in the development of T cell immunity [9,42][9][42]. DAF deficiency increases cytokine production by T cells, and CD59 ligation decreases CD4+ T cell activation [43]. The interaction between antigen-presenting cells (APC) and T lymphocytes induces the local production of C3, C5, as well as FB and FD. Moreover, C3aR and C5aR receptors are upregulated on T lymphocytes, whereas DAF production is downregulated. Local production of complement system components from immune cells allows signals to be transduced by C3aR and C5aR receptors in an autocrine and paracrine manner. Complement system component C3 activated in the alternative pathway can increase the production of proinflammatory cytokines from T cells [44,45][44][45]. Induction of Th1 responses also depends on the activation of C3aR and CD46 receptor on T cells through their T cell-derived ligands [46]. In contrast, the absence of C3aR and C5aR receptors leads to reduced complement protein and receptor regulation, lack of expression of co-stimulatory molecules, impaired production of cytokines (IL-1, IL-23, and IL-12), induction of the Treg cell response, and inhibition of T cell proliferation [47,48,49][47][48][49].
3. Antiviral Activity of the Complement System and Viral Strategies for Reducing the Complement System Action
All three complement system pathways can lead to viral opsonization and deposition of complement system components upon activation. The outcome of this response is highly dependent on the infectious agent and can enhance viral infection, suppress viral infection, or be dysregulated by the expression of certain viral proteins [50]. The MBL protein of the lectin pathway can interact with numerous viral antigens and have different effects on neutralization or increased viral replication. MBL can directly bind the glycoprotein (GP) of the Ebola virus (EBOV) [51]. High doses of MBL, relative to other complement proteins, can enhance infection with nonreplicating (pseudotyped) EBOV-GP virus in primary human macrophages and human monocyte-derived macrophage cell lines [51]. This may be due to the activation by viral ligands of the C1QBP (gC1qR) translocation, which inhibits RIG-1 and then inhibits antiviral signaling by downregulating type I interferons. It is speculated that EBOV-MBL complexes activate C1QBP, which then negatively regulates RIG-1 inhibition of viral infection, thereby enhancing viral proliferation [51]. Additionally, MBL opsonization of the EBOV GP prevents GP binding to DC-SIGN (dendritic cell-specific intercellular adhesion molecule) and, therefore, neutralizes EBOV (pseudotype) [52]. Thus, in the context of EBOV infection, MBL effects appear to be dependent on the cellular target and the relative concentrations of other complement system protein components. It has also been shown that in vitro infection by the human immunodeficiency virus (HIV) of CD4+ H9 lymphoblasts is inhibited by MBL from human serum. In addition, MBL is able to selectively bind to HIV-infected H9 cells and HIV-infected U937 cell line [53]. These results indicate that MBL inhibits viral entry to susceptible cells. Similarly, in another study [53], gp120 HIV bound directly to MBL. In a later study about HIV, MBL was shown to be sufficient for virus opsonization but not neutralization [54]. It also showed that both the primary isolates (PI) of HIV and cell line-adapted HIV, despite binding to MBL, are relatively resistant to neutralization by MBL at the levels normally present in the serum. However, binding and opsonization of HIV by MBL may alter virus trafficking and viral-antigen presentation during HIV infection [54]. In another study, ref. [55] complement opsonization of herpes simplex virus-2 (HSV-2) both by human serum and by seminal plasma, produced enhanced infection of DCs and resulted in greater productive infection compared to free, nonopsonized HSV-2. Furthermore, opsonization gave rise to significantly higher gene expression of all inflammatory (TNF-α, IL-6, IL-1β) and antiviral factors (IFN-α, IFN-β, MX1), but at the protein level these differences between free and complement-opsonized HSV-2 were not as clear as at the gene level. The enhanced infection induced by complement-opsonized virions required the functional complement receptor 3 (CR3). In contrast, the presence of complement in combination with HSV-1 or HSV-2-specific antibodies decreased infection, inflammation, and antiviral responses of DCs. HSV-2 infection of DCs required endocytosis and endosomal acidification, as inhibition of these cellular events decreased infection [55].
Other complement proteins and subsequently the complement system activation products can opsonize virus particles. In the case of dengue virus (DENV) and West Nile virus (WNV), viral neutralization occurs in a C3- and C4-dependent manner after MBL binding. In the case of WNV, neutralization was achieved despite reduced levels of C5, indicating that neutralization did not require MAC generation [56]. In the case of monkey simian virus 5 (SV5), complement-mediated neutralization is achieved mainly through C3 deposition and the formation of viral aggregates rather than viral lysis [57]. Similarly, the complement activation in the presence of Influenza A Virus (IVA) results in viral aggregation and opsonization of the hemagglutinin receptor, although IgM antibodies and activation of the classical pathway are required to achieve neutralization [58]. Some complement proteins may also have a protective intracellular function. Intracellular C3 signaling induces the production of proinflammatory cytokines (IFN-β, IL-6, and IL-1β) by nuclear factor kappa B (NF-κB), and activates interferon regulatory factor (IRF), and activator protein-1 (AP-1). Detection of intracellular C3 has been shown to be dependent on mitochondrial antiviral signaling protein (MAVS) and independent of PAMPs and pattern recognition receptors (PRRs) [59]. Infected host cells that present viral antigens on the cell surface membrane can activate the classical pathway as the antigens bind IgM/IgG, and induce complement-dependent cytotoxicity (CDC). The infected cell is then lysed by MAC to reduce the virus titer. However, some viruses have evolved self-defense mechanisms against the action of the complement system to enable their survival [50] (Table 2).
Table 2. Complement evasion strategies used by selected viruses.
| Disease | Virus | Family | Strategies Evasion | References |
|---|---|---|---|---|
| Influenza | Influenza viruses |
Orthomyxoviridae | Virus acquires CD59 on the surface and inhibits C1q-mediated recognition of virions Inhibition of neutralization by blocking the interaction of C1q with antibodies bound to the viral surface Inability of human C3b to recognize the surface of the virus and its opsonization |
[60][61][62] |
| Severe Acute Respiratory Syndrome (SARS) Middle East Respiratory Syndrome (MERS) Coronavirus disease (COVID-19) |
SARS-CoV MERS-CoV SARS-CoV-2 |
Coronaviridae | No data | - |
| Viral Lower Respiratory Tract Illiness |
Respiratory syncytial virus (RSV) |
Pneumoviridae | Transcriptional regulation of complement proteins |
[63] |
| Hepatitis B (HB) | Hepatitis B virus (HBV) |
Hepadnaviridae | The HBV X protein (HBx) upregulates CD59 and C4b-binding protein α (C4BPα), which inhibit the formation of MAC and provides protection from complement-mediated cytolysis | [64][65] |
| Ebola Virus Disease (EVD) | Ebola virus (EBOV) | Filoviridae | No data | - |
| Zika virus (ZIKV) | Incorporation into the viral envelope the of regulatory protein CD55 which contributes to virus stability and helps to avoid complement-dependent virolysis | [70] | ||
| West Nile Fever (WNF) | West Nile virus (WNV) |
NS1 directly binds and recruits FH to the surface of infected cells resulting in the inhibition of complement activation in all pathways and MAC formation | [71] |
References
- Mathern, D.R.; Heeger, P.S. Molecules Great and Small: The Complement System. Clin. J. Am. Soc. Nephrol. 2015, 10, 1636–1650.
- Walport, M.J. Complement. First of two parts. N. Engl. J. Med. 2001, 344, 1058–1066.
- Cochrane, C.G.; Unanue, E.R.; Dixon, F.J. A Role of Polymorphonuclear Leukocytes and Complement in Nephrotoxic Nephritis. J. Exp. Med. 1965, 122, 99–116.
- Mastellos, D.; Morikis, D.; Isaacs, S.N.; Holland, M.C.; Strey, C.W.; Lambris, J.D. Complement: Structure, functions, evolution, and viral molecular mimicry. Immunol. Res. 2003, 27, 367–386.
- Janeway, C.A., Jr.; Travers, P.; Walport, M.; Shlomchik, M.J. Immunobiology: The Immune System iN. Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001.
- Morgan, B. The Human Complement System in Health and Disease. Ann. Rheum. Dis. 1998, 57, 581.
- Cho, H. Complement regulation: Physiology and disease relevance. Korean J. Pediatr. 2015, 58, 239–244.
- Merle, N.S.; Noe, R.; Halbwachs-Mecarelli, L.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement System Part II: Role in Immunity. Front. Immunol. 2015, 6, 257.
- Dunkelberger, J.R.; Song, W.C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50.
- Gaboriaud, C.; Thielens, N.M.; Gregory, L.A.; Rossi, V.; Fontecilla-Camps, J.C.; Arlaud, G.J. Structure and activation of the C1 complex of complement: Unraveling the puzzle. Trends Immunol. 2004, 25, 368–373.
- Wallis, R.; Mitchell, D.A.; Schmid, R.; Schwaeble, W.J.; Keeble, A.H. Paths reunited: Initiation of the classical and lectin pathways of complement activation. Immunobiology 2010, 215, 1–11.
- Medzhitov, R.; Janeway, C., Jr. Innate immunity. N. Engl. J. Med. 2000, 343, 338–344.
- Medzhitov, R.; Janeway, C.A., Jr. Decoding the patterns of self and nonself by the innate immune system. Science 2002, 296, 298–300.
- Harmat, V.; Gal, P.; Kardos, J.; Szilagyi, K.; Ambrus, G.; Vegh, B.; Naray-Szabo, G.; Zavodszky, P. The structure of MBL-associated serine protease-2 reveals that identical substrate specificities of C1s and MASP-2 are realized through different sets of enzyme-substrate interactions. J. Mol. Biol. 2004, 342, 1533–1546.
- Gal, P.; Barna, L.; Kocsis, A.; Zavodszky, P. Serine proteases of the classical and lectin pathways: Similarities and differences. Immunobiology 2007, 212, 267–277.
- Gupta, P.; Tripathy, A.S. Alternative pathway of complement activation has a beneficial role against Chandipura virus infection. Med. Microbiol. Immunol. 2020, 209, 109–124.
- Walport, M.J. Complement. Second of two parts. N. Engl. J. Med. 2001, 344, 1140–1144.
- Hourcade, D.E. Properdin and complement activation: A fresh perspective. Curr. Drug. Targets 2008, 9, 158–164.
- Harboe, M.; Mollnes, T.E. The alternative complement pathway revisited. J. Cell Mol. Med. 2008, 12, 1074–1084.
- Muller-Eberhard, H.J. The killer molecule of complement. J. Investig. Dermatol. 1985, 85, 47s–52s.
- Wetsel, R.A.; Kildsgaard, J.; Haviland, D.L. Complement anaphylatoxins (C3a, C4a, C5a) and their receptors (C3aR, C5aR/CD88) as therapeutic targets in inflammation. In Therapeutic Interventions in the Complement System. Contemporary Immunology; Lambris, J.D., Holers, M., Eds.; Humana Press: Totowa, NJ, USA, 2000; pp. 113–153.
- Wang, H.; Ricklin, D.; Lambris, J.D. Complement-activation fragment C4a mediates effector functions by binding as untethered agonist to protease-activated receptors 1 and 4. Proc. Natl. Acad. Sci. USA 2017, 114, 10948–10953.
- Williams, T.J. Vascular permeability changes induced by complement-derived peptides. Agents Actions 1983, 13, 451–455.
- Takabayashi, T.; Vannier, E.; Clark, B.D.; Margolis, N.H.; Dinarello, C.A.; Burke, J.F.; Gelfand, J.A. A new biologic role for C3a and C3a desArg: Regulation of TNF-alpha and IL-1 beta synthesis. J. Immunol. 1996, 156, 3455–3460.
- Monsinjon, T.; Gasque, P.; Chan, P.; Ischenko, A.; Brady, J.J.; Fontaine, M.C. Regulation by complement C3a and C5a anaphylatoxins of cytokine production in human umbilical vein endothelial cells. FASEB J. 2003, 17, 1003–1014.
- Atkinson, J.P.; Du Clos, T.W.; Mold, C.; Kulkarni, H.; Hourcade, D.; Wu, X. The Human Complement System: Basic Concepts and Clinical Relevance. Capture 21; Elsevier: Amsterdam, The Netherlands, 2019.
- Zipfel, P.F.; Skerka, C. Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 2009, 9, 729–740.
- Liszewski, M.K.; Farries, T.C.; Lublin, D.M.; Rooney, I.A.; Atkinson, J.P. Control of the complement system. Adv. Immunol. 1996, 61, 201–283.
- Giannakis, E.; Jokiranta, T.S.; Male, D.A.; Ranganathan, S.; Ormsby, R.J.; Fischetti, V.A.; Mold, C.; Gordon, D.L. A common site within factor H SCR 7 responsible for binding heparin, C-reactive protein and streptococcal M protein. Eur. J. Immunol. 2003, 33, 962–969.
- Manuelian, T.; Hellwage, J.; Meri, S.; Caprioli, J.; Noris, M.; Heinen, S.; Jozsi, M.; Neumann, H.P.; Remuzzi, G.; Zipfel, P.F. Mutations in factor H reduce binding affinity to C3b and heparin and surface attachment to endothelial cells in hemolytic uremic syndrome. J. Clin. Investig. 2003, 111, 1181–1190.
- Morgan, H.P.; Schmidt, C.Q.; Guariento, M.; Blaum, B.S.; Gillespie, D.; Herbert, A.P.; Kavanagh, D.; Mertens, H.D.; Svergun, D.I.; Johansson, C.M.; et al. Structural basis for engagement by complement factor H of C3b on a self surface. Nat. Struct. Mol. Biol. 2011, 18, 463–470.
- Kim, D.D.; Song, W.C. Membrane complement regulatory proteins. Clin. Immunol. 2006, 118, 127–136.
- Kemper, C.; Atkinson, J.P.; Hourcade, D.E. Properdin: Emerging roles of a pattern-recognition molecule. Annu. Rev. Immunol. 2010, 28, 131–155.
- Agarwal, S.; Ferreira, V.P.; Cortes, C.; Pangburn, M.K.; Rice, P.A.; Ram, S. An evaluation of the role of properdin in alternative pathway activation on Neisseria meningitidis and Neisseria gonorrhoeae. J. Immunol. 2010, 185, 507–516.
- Carroll, M.C. The complement system in B cell regulation. Mol. Immunol. 2004, 41, 141–146.
- Fang, Y.; Xu, C.; Fu, Y.X.; Holers, V.M.; Molina, H. Expression of complement receptors 1 and 2 on follicular dendritic cells is necessary for the generation of a strong antigen-specific IgG response. J. Immunol. 1998, 160, 5273–5279.
- Carter, R.H.; Fearon, D.T. CD19: Lowering the threshold for antigen receptor stimulation of B lymphocytes. Science 1992, 256, 105–107.
- Barrington, R.A.; Zhang, M.; Zhong, X.; Jonsson, H.; Holodick, N.; Cherukuri, A.; Pierce, S.K.; Rothstein, T.L.; Carroll, M.C. CD21/CD19 coreceptor signaling promotes B cell survival during primary immune responses. J. Immunol. 2005, 175, 2859–2867.
- Fischer, M.B.; Goerg, S.; Shen, L.; Prodeus, A.P.; Goodnow, C.C.; Kelsoe, G.; Carroll, M.C. Dependence of germinal center B cells on expression of CD21/CD35 for survival. Science 1998, 280, 582–585.
- Fischer, W.H.; Hugli, T.E. Regulation of B cell functions by C3a and C3a(desArg): Suppression of TNF-alpha, IL-6, and the polyclonal immune response. J. Immunol. 1997, 159, 4279–4286.
- Ottonello, L.; Corcione, A.; Tortolina, G.; Airoldi, I.; Albesiano, E.; Favre, A.; D’Agostino, R.; Malavasi, F.; Pistoia, V.; Dallegri, F. rC5a directs the in vitro migration of human memory and naive tonsillar B lymphocytes: Implications for B cell trafficking in secondary lymphoid tissues. J. Immunol. 1999, 162, 6510–6517.
- Longhi, M.P.; Harris, C.L.; Morgan, B.P.; Gallimore, A. Holding T cells in check—A new role for complement regulators? Trends Immunol. 2006, 27, 102–108.
- Le Friec, G.; Kemper, C. Complement: Coming full circle. Arch. Immunol. Ther. Exp. 2009, 57, 393–407.
- Liszewski, M.K.; Kolev, M.; Le Friec, G.; Leung, M.; Bertram, P.G.; Fara, A.F.; Subias, M.; Pickering, M.C.; Drouet, C.; Meri, S.; et al. Intracellular complement activation sustains T cell homeostasis and mediates effector differentiation. Immunity 2013, 39, 1143–1157.
- Elvington, M.; Liszewski, M.K.; Bertram, P.; Kulkarni, H.S.; Atkinson, J.P. A C3(H20) recycling pathway is a component of the intracellular complement system. J. Clin. Investig. 2017, 127, 970–981.
- Ghannam, A.; Fauquert, J.L.; Thomas, C.; Kemper, C.; Drouet, C. Human complement C3 deficiency: Th1 induction requires T cell-derived complement C3a and CD46 activation. Mol. Immunol. 2014, 58, 98–107.
- Heeger, P.S.; Lalli, P.N.; Lin, F.; Valujskikh, A.; Liu, J.; Muqim, N.; Xu, Y.; Medof, M.E. Decay-accelerating factor modulates induction of T cell immunity. J. Exp. Med. 2005, 201, 1523–1530.
- Strainic, M.G.; Liu, J.; Huang, D.; An, F.; Lalli, P.N.; Muqim, N.; Shapiro, V.S.; Dubyak, G.R.; Heeger, P.S.; Medof, M.E. Locally produced complement fragments C5a and C3a provide both costimulatory and survival signals to naive CD4+ T cells. Immunity 2008, 28, 425–435.
- Strainic, M.G.; Shevach, E.M.; An, F.; Lin, F.; Medof, M.E. Absence of signaling into CD4(+) cells via C3aR and C5aR enables autoinductive TGF-beta1 signaling and induction of Foxp3(+) regulatory T cells. Nat. Immunol. 2013, 14, 162–171.
- Mellors, J.; Tipton, T.; Longet, S.; Carroll, M. Viral Evasion of the Complement System and Its Importance for Vaccines and Therapeutics. Front. Immunol. 2020, 11, 1450.
- Brudner, M.; Karpel, M.; Lear, C.; Chen, L.; Yantosca, L.M.; Scully, C.; Sarraju, A.; Sokolovska, A.; Zariffard, M.R.; Eisen, D.P.; et al. Lectin-dependent enhancement of Ebola virus infection via soluble and transmembrane C-type lectin receptors. PLoS ONE 2013, 8, e60838.
- Ji, X.; Olinger, G.G.; Aris, S.; Chen, Y.; Gewurz, H.; Spear, G.T. Mannose-binding lectin binds to Ebola and Marburg envelope glycoproteins, resulting in blocking of virus interaction with DC-SIGN and complement-mediated virus neutralization. J. Gen. Virol. 2005, 86, 2535–2542.
- Ezekowitz, R.A.; Kuhlman, M.; Groopman, J.E.; Byrn, R.A. A human serum mannose-binding protein inhibits in vitro infection by the human immunodeficiency virus. J. Exp. Med. 1989, 169, 185–196.
- Ying, H.; Ji, X.; Hart, M.L.; Gupta, K.; Saifuddin, M.; Zariffard, M.R.; Spear, G.T. Interaction of mannose-binding lectin with HIV type 1 is sufficient for virus opsonization but not neutralization. AIDS Res. Hum. Retrovir. 2004, 20, 327–335.
- Crisci, E.; Ellegard, R.; Nystrom, S.; Rondahl, E.; Serrander, L.; Bergstrom, T.; Sjowall, C.; Eriksson, K.; Larsson, M. Complement Opsonization Promotes Herpes Simplex Virus 2 Infection of Human Dendritic Cells. J. Virol. 2016, 90, 4939–4950.
- Fuchs, A.; Lin, T.Y.; Beasley, D.W.; Stover, C.M.; Schwaeble, W.J.; Pierson, T.C.; Diamond, M.S. Direct complement restriction of flavivirus infection requires glycan recognition by mannose-binding lectin. Cell Host Microbe 2010, 8, 186–195.
- Johnson, J.B.; Capraro, G.A.; Parks, G.D. Differential mechanisms of complement-mediated neutralization of the closely related paramyxoviruses simian virus 5 and mumps virus. Virology 2008, 376, 112–123.
- Jayasekera, J.P.; Moseman, E.A.; Carroll, M.C. Natural antibody and complement mediate neutralization of influenza virus in the absence of prior immunity. J. Virol. 2007, 81, 3487–3494.
- Tam, J.C.; Bidgood, S.R.; McEwan, W.A.; James, L.C. Intracellular sensing of complement C3 activates cell autonomous immunity. Science 2014, 345, 1256070.
- Shaw, M.L.; Stone, K.L.; Colangelo, C.M.; Gulcicek, E.E.; Palese, P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008, 4, e1000085.
- Rattan, A.; Pawar, S.D.; Nawadkar, R.; Kulkarni, N.; Lal, G.; Mullick, J.; Sahu, A. Synergy between the classical and alternative pathways of complement is essential for conferring effective protection against the pandemic influenza A(H1N1) 2009 virus infection. PLoS Pathog. 2017, 13, e1006248.
- Zhang, J.; Li, G.; Liu, X.; Wang, Z.; Liu, W.; Ye, X. Influenza A virus M1 blocks the classical complement pathway through interacting with C1qA. J. Gen. Virol. 2009, 90, 2751–2758.
- Kumar, N.A.; Kunnakkadan, U.; Thomas, S.; Johnson, J.B. In the Crosshairs: RNA Viruses OR Complement? Front. Immunol. 2020, 11, 573583.
- Shan, C.; Zhang, S.; Cui, W.; You, X.; Kong, G.; Du, Y.; Qiu, L.; Ye, L.; Zhang, X. Hepatitis B virus X protein activates CD59 involving DNA binding and let-7i in protection of hepatoma and hepatic cells from complement attack. Carcinogenesis 2011, 32, 1190–1197.
- Feng, G.; Li, J.; Zheng, M.; Yang, Z.; Liu, Y.; Zhang, S.; Ye, L.; Zhang, W.; Zhang, X. Hepatitis B virus X protein up-regulates C4b-binding protein alpha through activating transcription factor Sp1 in protection of hepatoma cells from complement attack. Oncotarget 2016, 7, 28013–28026.
- Avirutnan, P.; Hauhart, R.E.; Somnuke, P.; Blom, A.M.; Diamond, M.S.; Atkinson, J.P. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J. Immunol. 2011, 187, 424–433.
- Thiemmeca, S.; Tamdet, C.; Punyadee, N.; Prommool, T.; Songjaeng, A.; Noisakran, S.; Puttikhunt, C.; Atkinson, J.P.; Diamond, M.S.; Ponlawat, A.; et al. Secreted NS1 Protects Dengue Virus from Mannose-Binding Lectin-Mediated Neutralization. J. Immunol. 2016, 197, 4053–4065.
- Kurosu, T.; Chaichana, P.; Yamate, M.; Anantapreecha, S.; Ikuta, K. Secreted complement regulatory protein clusterin interacts with dengue virus nonstructural protein 1. Biochem. Biophys. Res. Commun. 2007, 362, 1051–1056.
- Conde, J.N.; da Silva, E.M.; Allonso, D.; Coelho, D.R.; Andrade, I.D.S.; de Medeiros, L.N.; Menezes, J.L.; Barbosa, A.S.; Mohana-Borges, R. Inhibition of the Membrane Attack Complex by Dengue Virus NS1 through Interaction with Vitronectin and Terminal Complement Proteins. J. Virol. 2016, 90, 9570–9581.
- Malekshahi, Z.; Bernklau, S.; Schiela, B.; Koske, I.; Banki, Z.; Stiasny, K.; Harris, C.L.; Wurzner, R.; Stoiber, H. Incorporation of CD55 into the Zika Viral Envelope Contributes to Its Stability against Human Complement. Viruses 2021, 13, 510.
- Chung, K.M.; Liszewski, M.K.; Nybakken, G.; Davis, A.E.; Townsend, R.R.; Fremont, D.H.; Atkinson, J.P.; Diamond, M.S. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc. Natl. Acad. Sci. USA 2006, 103, 19111–19116.
More
