Tantisira conducted a GWAS in 2011 and identified the glucocorticoid-induced transcript 1 (GLCCI1) gene and the SNPs, rs37972 and rs37973 (
Table 15)
[12][51]. Non-Hispanic white children selected from the CAMP study were given budesonide, and the response was measured by the change in FEV1 from baseline. Although little is known about GLCCI1, those homozygote for the mutant T allele at SNP, rs37972, yielded a poorer response compared to those who were heterozygote (CT) or homozygote (CC) (OR, 2.36; 95% CI, 1.27–4.41)
[12][51]. Additionally, linkage disequilibrium was found with the SNP, rs37973 (r
2 = 0.99). Individuals that were homozygote for the wild-type A allele had a better response to budesonide compared to those homozygotes for the mutant G allele (3.2 ± 1.6% vs. 9.4 ± 1.1% increase in FEV1)
[12][51]. Similar findings were seen in a trial conducted by Thompson et al.
[13][52]. Included in that study were 402 European children on long-term ICS treatment (>6 months). Higher steroid doses and increased hospitalization were seen in SNP, rs37973, homozygous for the mutant allele compared to those heterozygous or homozygous wild-type
[13][52]. Unlike Tantisira
[12][51], Vijverberg saw no association with individuals having SNP, rs37972 and ICS response in Northern European children
[14][53]. Additionally, Vijverberg utilized hospital visits and oral steroid use as markers for exacerbation and concluded that the mutant T allele was not associated with an increased risk when treated with budesonide
[14][53]. In contrast, Huang et al. found associations in the GLCCI1 gene in 263 Chinese children. SNPs, rs37969 GG, rs37972 CC, and rs37973 AA produced favorable changes measured by maximal mid-expiratory flow (
p ≤ 0.05)
[15][54]. Stress-induced phosphoprotein 1 (STIP1) was also explored
in th
ereis study and was shown to suppress inflammatory mediators. Although a significant association between SNP rs2236647 CC and the risk of developing asthma was found, there was no association with any SNP and ICS response
[15][54].
Table 15.
ICS and genetic variations.
| Study |
Population |
Gene |
Medication |
Measurement |
Results |
| Tantisira [12] | Tantisira [51] |
White |
GLCCI1 |
Budesonide |
Change in FEV1 from baseline |
rs37972 TT and rs37973 G allele variants showed unfavorable responses |
| Thompson [13] | Thompson [52] |
European |
GLCCI1 |
ICS |
Exacerbation (hospital visits and oral steroid use) |
rs37973 G allele showed unfavorable response |
| Vijverberg [14] | Vijverberg [53] |
Europe |
GLCCI1 |
Budesonide |
Exacerbation (emergency room visits, hospital visits, oral steroid use) |
No association found at rs37972 |
| Huang [15] | Huang [54] |
Chinese |
GLCCI1 |
ICS |
MMEF |
rs37969, rs37972, and rs37973 produced favorable response. |
| Kim [16] | Kim [55] |
Korean |
HDAC1 |
ICS |
% change in FEV1 pre- and post-bronchodilator |
rs1741981 CT and TT produced favorable response |
| Tantisira [17] | Tantisira [56] |
Caucasians |
CRHR1 |
Budesonide |
% change in FEV1 from baseline |
rs242941T produced favorable response |
| Tantisira [18] | Tantisira [57] |
Multiethnic |
TBX21 |
Budesonide |
% change in FEV1 and PC20 |
rs2240017Q produced favorable response |
| Tantisira [19] | Tantisira [58] |
Multiethnic |
FCER2 |
Budesonide |
Exacerbations (ER visits or hospitalization) |
rs28364072 CC produced unfavorable response |
| Koster [20] | Koster [59] |
European |
FCER2 |
ICS, ICS + salmeterol, ICS + salmeterol + montelukast |
Exacerbations (hospital visits and/or oral steroid use) |
rs28364072 CC produced unfavorable response |
| Keskin [21] | Keskin [60] |
Turkey |
NR3C1 |
Fluticasone |
FEV1 improvement at 4 h |
rs41423247 GG produced favorable response |
| Stockman [22] | Stockman [61] |
Whites |
CYP3A4, CYP3A5, and CYP3A7 |
Fluticasone |
Asthma control scores |
CYP3A4 *22 produced favorable response |
| Stockman [23] | Stockman [62] |
White |
CYP3A4, CYP3A5, and CYP3A7 |
Beclomethasone |
Asthma control scores |
CYP3A5 *3/*3 produced favorable response |
It was hypothesized that histone deacetylase I and 2 (HDAC1, HDAC2) play a positive role in airway responsiveness and inflammation. Kim studied 35 adults and 70 children with the HDAC1 SNP, rs1741981
[16][55]. Bronchodilation was measured as a % change in FEV1 from baseline
[16][55]. Adults with the CC genotype produced diminished responses compared to the CT or TT genotypes when administered systemic corticosteroids (
p = 0.018). The same was true for the children given inhaled corticosteroids. Those with the CC genotype had a FEV1 of 14.1% compared to either the CT or TT (19.4%,
p = 0.035).
The corticotropin-release hormone (CRH) is a known mediator to stress and its corresponding receptor, corticotropin-releasing hormone receptor 1 (CRHR1), is hypothesized to have a role in ICS response
[17][56]. In comparison to the unfavorable responses seen in the GLCCI1 variants, Tantisira observed an increase in response in those with the CRHR1 variants among three cohorts of both children and adults treated with an ICS. Response was measured by dFEV1 over a span of eight weeks in the CAMP replication study. Those with the rs242941 mutant TT genotypes displayed a change of 17.80 + 6.77 in FEV1 compared to a change of 7.57 + 1.50 in those with the wild-type CC. This was not seen in the initial adult study treated with flunisolide. SNP, rs1876828 variant, was also associated with triamcinolone responses in the adult replication study but was statistically insignificant in children
[15][54].
Variants in the T-box transcription factor (TBX21), which encodes for transcription factor T-bet, also displayed improvements in ICS response. Children with glutamine at amino acid position 33 in place of histidine (rs2240017) displayed better PC20
[18][57]. PC20 is the provocative concentration resulting in a 20% drop of FEV1 post methacholine challenge. After four years, PC20 for the 33Q individuals given budesonide was measured to be 27.7 mg/mL, placing them in the non-asthmatic range
[18][57]. Tantisira also conducted a study with a focus on the Fc fragment of IgE low affinity II receptor (FCER2) gene
[19][58]. FCER2 encodes for CD23, causing IgE mediated responses which have been associated with severe asthmatic exacerbations. In this study, 311 Caucasian children were assigned to inhaled budesonide and observed over four years. The SNPs identified to be associated with increased IgE levels and exacerbations were: rs4996974, rs7249320, and rs28364072. Children with the SNP, rs28364072, homozygous for the mutant C allele were at a higher risk of severe exacerbations in both African American and Caucasian populations (HR 3.08 and 3.95, respectively)
[19][58]. Koster replicated these findings in two pediatric cohorts treated with ICS. Children’s homozygotes for the variant C allele had a higher risk of severe exacerbations (OR 2.38, 95% CI 1.47–3.85,
p = 0.0004), increase in hospital visits (OR 1.91, 95% CI 1.08–3.40,
p = 0.03), and were more likely to use higher ICS doses (OR 2.46, 95% CI 1.38–4.39,
p = 0.002
[20][59]. These findings support the need for earlier detection of FCER2 polymorphisms to identify those resistant to steroid treatment and provide timely anti-IgE treatment.
Located on chromosome 5q31-q32, nuclear receptor subfamily 3, group C, member 1 (NR3C1) is a protein-coding gene for glucocorticoid receptors. Keskin studied 82 children with a mean age of 9.6, given inhaled fluticasone propionate specifically evaluating the role of NR3C1
[21][60]. Of the 82 children, 26 had the rs41423247, GG genotype and displayed a better response compared to those with the CG or CC variations, measured by FEV1 (24.2% vs. 7.9%,
p = 0.006)
[21][60]. Stockman also conducted a study involving 734 Caucasian children receiving inhaled fluticasone propionate. Variations among cytochrome p450 (CYP) 3A4, 3A5, and 3A7, which are involved in fluticasone metabolism, were assessed for symptom control using the asthma control score. CYP3A5 and CYP3A7 were not found to have an association, however, CYP3A4 *22 children displayed better asthma control due to the reduced activity of the metabolic enzyme, leading to increased therapeutic outcomes
[22][61]. These findings were not seen in a follow-up study of 64 children treated with beclomethasone
[23][62]. They observed that children with CYP3A5 *3/*3, commonly found among Caucasian populations, displayed better asthma control, compared to either *1/*1 or *1/*3 variations
[23][62]. Pharmacogenomics testing on these CYP enzymes has the potential to provide guidance on future regimens. Children with the CYP3A4 *1/*1 or *1/*3 variation may be advised to use a non-beclomethasone medication, such as budesonide, as their initial ICS. Similarly, those with the CYP3A4 *22 variation may be advised to start therapy with fluticasone.
3. Leukotriene Modifiers (LTM)
Leukotrienes are involved in inflammatory processes and bronchoconstriction, both of which are implicated in asthmatic symptoms. Leukotriene modifiers are considered second line after ICS and LABA for chronic asthma according to the GINA guidelines
[3]. Administered orally, these LTMs include zileuton, montelukast, and zafirlukast which are subsequently classified into leukotriene synthesis inhibitors or leukotriene receptor antagonists. Zileuton inhibits the 5-lipoxygenase (5-LO) enzyme, halting the conversion of arachidonic acid into LTA4. Montelukast, zafirlukast, and pranlukast are cysteinyl-leukotriene receptor (CysLTR) inhibitors, antagonizing the effects of LTC4, LTD4, and LTE4
[24][25][63,64].
Similar to bronchodilators and ICS, limitations of leukotriene modifiers are highlighted by genetic variations, emphasizing inter-patient variability leading to exacerbated symptoms
[26][27][65,66]. In addition, nearly half of the side effects in children taking a LTM are attributed to psychiatric disturbances such as hallucinations and agitation
[25][64]. Genes such as arachidonate 5-lipoxygenase (ALOX5) and cysteinyl-LTs (CysLTs) previously identified and replicated in adult studies have not yet been fully confirmed in children. Identifying variants associated with positive leukotriene modifier response can further benefit children who cannot gain symptom control with ICS or beta agonists.
3.1. Genes That Affect Montelukast Response
Montelukast, favored in the pediatric community for its once-a-day dosing, is an oral medication for chronic asthma and exercise-induced bronchoconstriction (EIB). It binds to the CysT1 receptor (CysLTR1), antagonizing the effects of LTD4, and decreasing bronchoconstriction
[27][66].
Both pharmacodynamic and pharmacokinetic studies have shown to potentially impact montelukast responsiveness in children (
Table 26).
Table 26.
Montelukast and genetic variations.
| Study |
Population |
Gene |
Measurement |
Results |
| Kim [28] | Kim [67] |
Korean |
TBXA2 |
FEV1 |
Combination of +795 CT/CC and +924 TT showed unfavorable response |
| Klotsman [29] | Klotsman [69] |
Multiethnic |
ALOX5
CystLTR2
LTC4S |
PEF
PEF
FEV1 and PEF |
rs4987105 TT and rs4986832 AA variants showed favorable outcomes
rs912277 TT and rs912278 TC variants produced favorable response
No association found at rs730012 |
| Telleria [30] | Telleria [70] |
Spain |
ALOX5 |
% change in FEV1, exacerbations, and rescue inhaler need |
rs59439148 5/5 and 5/4 copies showed favorable outcomes |
| Kang [31] | Kang [71] |
Korean |
LTC4S
PTGDR |
≥10% increase in FEV1 post exercise challenge |
No significance at rs730012
rs803010 TT produced favorable response |
| Lee [32] | Lee [72] |
Korean |
LTC4S
CystLTR1 |
>10% increase in FEV1 post exercise challenge |
No significance at SNP rs730012
No significance found |
| Whelan [33] | Whelan [73] |
African American and Caucasian |
LTC4S |
FENO |
rs730012 AC produced favorable response |
| Mougey [34] | Mougey [74] |
Multiethnic |
SLCO2B1 |
Asthma symptom utility index (ASUI) |
rs12422149 GG produced favorable response
No association at rs2306168 |
| Li [35] | Li [75] |
Chinese |
SLCO2B1CYP2C8 |
Drug clearance |
Decreased clearance in rs12422149 GG No association found |
Thromboxane A2 (TBXA2), an arachidonic acid derivative, binds to the thromboxane A2 receptor (TBXA2R) and causes pulmonary smooth muscle constriction and platelet aggregation. To determine an association between the TBXA2R gene and LTM response, Kim et al. conducted a study on 695 Korean children with exercise-induced bronchospasms given montelukast. They found that those with the TBXA2R +795 CC or CT variations were 2.5 times more likely to be unresponsive to treatment (
p = 0.063)
[28][67]. Responders were defined as >10% in FEV1 after montelukast treatment. In addition, those with both the TBXA2R +795 CT or CC and +924 TT variation were less likely to respond to montelukast treatment (OR 3.67,
p = 0.041)
[28][67].
In comparison, Klotsman found a statistically significant association of responsiveness in the ALOX5 variants among children 15 years of age and older. ALOX5 encodes for 5-lipoxygenase (5-LO) and is found on chromosome 10q11.21
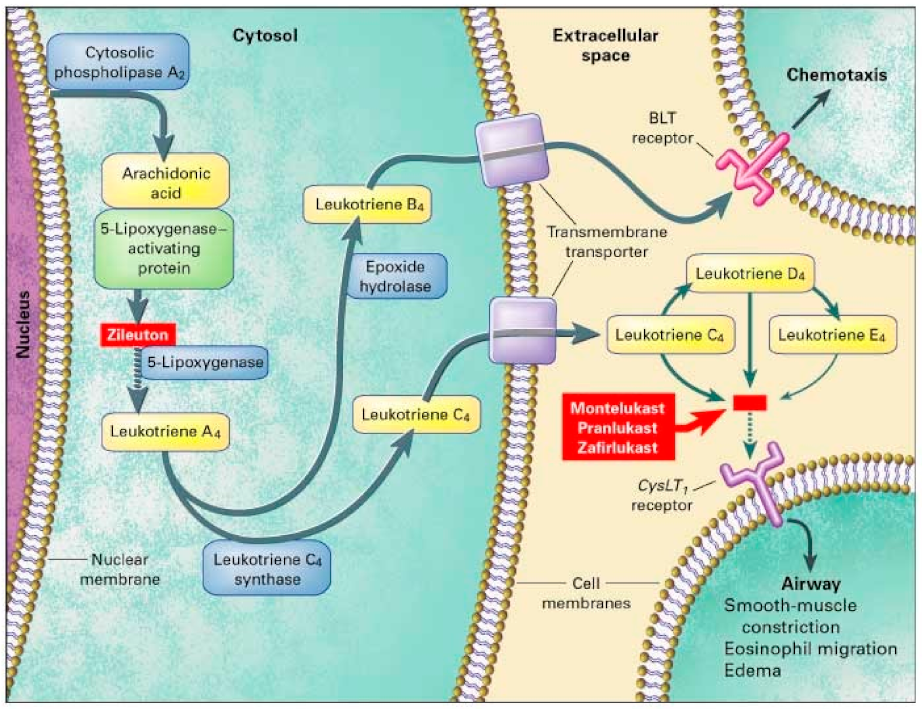
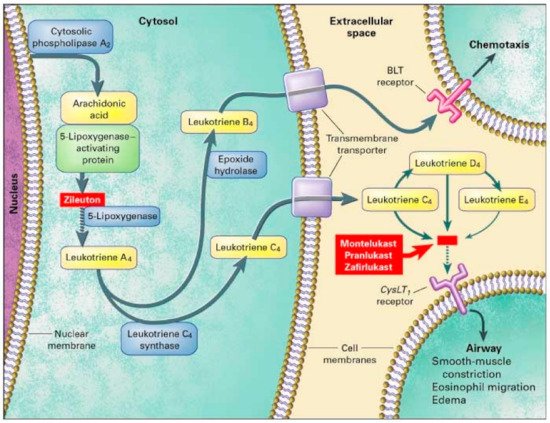
[36][68]. 5-LO is involved in the conversion of arachidonic acid into LTA4 and is the rate limiting step in leukotriene synthesis (
Figure 23). Two out of five variants were significantly associated with a positive response to montelukast. Those with the SNP rs4987105 TT genotype produced a mean PEF of 94.8 while those with CC genotypes were 33.7 (
p = 0.01). Those with the SNP rs4986832 AA genotype produced a mean PEF of 102.4 while those with the GG genotype were 34.9 (
p = 0.01)
[29][69]. Conversely, Telleria et al. studied the promoter region of the ALOX5 gene. Five copies of the transcription factor binding sequence GGGCGG (rs59439148) were recognized as the major allele, with variants hypothesized to produce reduced activity
[30][70]. They found greater improvements with homozygotes wild type (5/5) and heterozygotes (5/4), compared to homozygote variants (4/4) after 6 months of montelukast treatment. Those with the 4/4 repeats displayed higher rates of exacerbations, worsening FEV1, and greater need for rescue inhaler compared to those with the 5/5 or 5/4 repeats (
p ≤ 0.05)
[30][70].
Figure 23. Leukotriene Modifiers Mechanism of Action. Adapted from [36]. Leukotriene Modifiers Mechanism of Action. Adapted from [68].
Similar to CystLTR1, cysteinyl- leukotriene (CystLT) receptor 2 (CysLTR2) have recently been found on smooth muscle cells. Although the variant function of this gene has yet to be identified, it has been associated with asthmatic treatment response. SNPs, rs912277 TC and rs912278 CC, were found to produce greater PEF (
p = 0.021 and
p = 0.02, respectively) in a study done by Klotsman
[29][69]. Participants were at least 15 years of age and observed over 12 weeks. TT genotypes in both SNPs produced a mean PEF of 38.8 and 29.1, respectively. In the same study, they assessed leukotriene C4 synthase (LTC4S). LTC4S converts leukotriene A4 to leukotriene C4, increasing CysLT levels contributing to asthmatic inflammation (
Figure 23). However, no association was found in montelukast treatment in either FEV1 or PEF in SNP, rs730012
[29][69]. Furthermore, Kang could not establish statistical significance in those with the LTC4S polymorphisms between responders and non-responders (
p = 0.702) in 100 Korean asthmatic children
[31][71]. Lee claimed that montelukast responsiveness is associated with total IgE and PC20 levels, opposed to FEV1
[32][72]. This
res
earchtudy also observed CysLTR1 variations with exercise-induced bronchoconstriction (EIB). However, no correlation to montelukast responsiveness, measured by >10% increase in FEV1 post exercise challenge, was found
[32][72]. Whelan investigated 13 children, ages 10–16 and found a positive correlation in montelukast responsiveness and LTC4S. Statistical significance in those with the LTC4S AC genotype produced a greater decrease in fraction of exhaled nitric oxide (FENO) (slope −3.13%/day), which was correlated with a decrease in airway inflammation. Those with the AA genotype did not produce any noticeable change in FENO, suggesting montelukast use in these patients may not be beneficial
[33][73].
A study by Kang on 10 Korean asthmatic children investigated prostaglandin D2 receptor (PTGDR), a derivative of arachidonic acid implicated in eosinophilia migration
[31][71]. Over the span of eight weeks children receiving montelukast with the rs803010 TT genotype had a greater FEV1 fall post-exercise challenge compared to those with the TC or CC genotype (63.8% vs. 36.2%
p = 0.038). No difference in response was found when analyzing both PTGDR and LTC4S variants
[31][71].
Lipworth observed 62 children with ADRB2 Arg16Arg variants and found montelukast reduced school absences and symptoms compared to those treated with salmeterol. All children also received inhaled fluticasone throughout the one-year study. It was concluded that the use of montelukast and an ICS may be more beneficial than the recommended ICS and LABA combination
[37][47] (
Table 37).
Table 37.
Montelukast Combination and ADRB2.
| Study |
Population |
Gene |
Medication |
Measurement |
Results |
| Lipworth [37] | Lipworth [47] |
Scotland |
ADRB2 |
Fluticasone + montelukast or salmeterol + fluticasone |
School absences, FEV1, asthma symptoms |
Arg16Arg produced favorable responses to fluticasone and montelukast |
Organic anion transporter polypeptide 2B1 (OATP2B1) are drug transporters found in the liver, intestine, and kidney and are responsible for reuptake of various substrates. This transporter is encoded by the gene solute carrier organic anion transporter family member 2B1 (SLCO2B1). OATP2B1 SNPs rs12422149 and rs2306168 have been associated with montelukast absorption
[34][74]. Mougey utilized the Asthma Symptoms Utility Index (ASUI) to measure drug responsiveness and found that those with the rs12422149 GG genotype demonstrated greater improvement at three months and six months compared to the those with the AG genotype
[34][74]. Similarly, a study by Li with 50 Chinese children with an average age of 4.4 years old found lower montelukast clearance in the SLCO2B1 rs12422149 GG genotype compared to the GA and AA (0.77 ± 0.21 vs. 0.94 ± 0.26,
p = 0.020)
[35][75]. Those with the GG genotype displayed better responsiveness to montelukast due to lower drug clearance leading to increased plasma concentrations. Li also examined CYP2C8 variations, however no association was found with montelukast clearance in Chinese children
[35][75]. These results may favor genetic testing to identify SLCO2B1, rs12422149 GG variants, which presumably should see the greatest effect from monteluakast treatment among LTM medications
[38][76]. Those without the SLCO2B1, rs12422149 GG variants may see better efficacy on a different LTM
[39][77].
3.2. Genes Affecting Other Leukotriene Modifier Responses
Zileuton is a 5-LO inhibitor that leads to decreased leukotriene synthesis, inflammation, and bronchoconstriction (
Figure 23). Those on zileuton should monitor their ALT regularly
[40][78]. Data on zileuton responsiveness is scarce as most studies utilized montelukast as the drug of choice. Tcheurekdjlan looked at Puerto Rican and Mexican youths with an average age of 12.3 taking a leukotriene modifier and albuterol. Bronchodilator responsiveness, measured by FEV1, was found to be greater in those taking LTM compared to those did not (
p = 0.001)
[41][79]. In addition, leukotriene A4 hydrolase (LTA4H) was investigated. LTA4H converts LTA4 to LTB4, which contributes to neutrophilic asthma characterized by severe airway obstruction. The LTA4H SNP rs2540491 minor A allele produced a change of 7–10% in FEV1 compared to the major G allele which produced a change of −0.31% in FEV1 (
Table 48). These results were seen in the Puerto Rican population, but not in the Mexican population. The LTA4H SNP rs2540487 GA heterozygotes also produced a change of more than 10% in FEV1 (
p < 0.001) compared to homozygotes major or minor (2.5 % change in FEV1,
p = 0.180 and 0.679, respectively)
[41][79]. Arachidonate 5-lipoxygenase activating protein (ALOX5AP) modulates downstream leukotriene synthesis along with ALOX5, converting arachidonic acid into leukotriene A4 (
Figure 23). No association was found between ALOX5AP, rs10507391 and rs955196 variants with leukotriene modifier bronchodilation. Interactions between LTA4H, ALOX5AP, and their variants were also analyzed for bronchodilation. The presence of ALOX5AP, rs10507391 major A allele and LTA4H minor allele variants contributed to bronchodilation, whereas the presence of ALOX5AP, rs9551963 minor C allele and LT4AH minor allele variants contributed to bronchodilation. These findings were significant in Puerto Rican populations but not Mexican populations
[41][79].
Table 48.
Leukotriene Modifier and Genetic Variants.
| Study |
Population |
Gene |
Medication |
Measurement |
Results |
| Tcheurekdjlan [41] | Tcheurekdjlan [79] |
Mexican and Puerto Ricans |
LTA4H
ALOX5AP |
Albuterol + leukotriene modifiers (montelukast, zafirlukast, and zileuton) |
% change in FEV1 pre and post bronchodilator |
LTA4H rs2540491 A variants and
rs2540487 GA variants produced favorable response
No association of ALOX5AP variants alone
rs10507391 A allele and rs9551963 C allele magnifies LTA4H response |