Bioluminescence imaging (BLI) is based on the catalytic activity of luciferase enzymes, which oxidize the substrate called luciferin to generate an excited-state molecule that emits bioluminescence.
- luciferase
- luciferin
- BLI
1. Introduction
In vivo bioluminescence imaging (BLI) has enabled visualization of biological processes in intact living organisms, providing abundant quantitative, spatiotemporal information that is far beyond the reach of conventional in vitro assays. Unlike the fluorescence imaging technique, BLI requires no external excitation light that might cause background fluorescence or phototoxicity in analyzed samples. Therefore, one can achieve exquisitely sensitive imaging in opaque heterogeneous tissues. Consequently, BLI has enabled in vivo monitoring of various biological processes such as tumor growth, cancer metastasis, and bacterial infection[1] [1].
2. Luciferases used in BLI
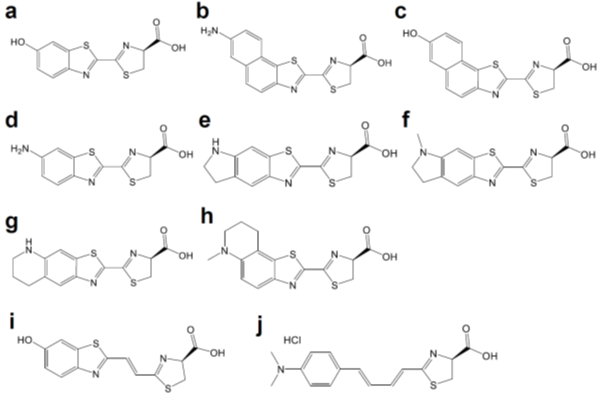
Luciferases used in BLI are derived from diverse species that include beetles, sea pansy, copepods, and deep-sea shrimp. The corresponding luciferins hold distinct chemical structures: firefly luciferase (FLuc, Figure 1a) requires d-luciferin (Figure 2a), while marine Renilla luciferase (RLuc, Figure 1b) and Oplophorus luciferase (OLuc) use coelenterazine (Figure 3b). All luciferins require chemical reactions to emit bioluminescence: the oxidation of luciferins by luciferases to yield excited-state oxyluciferin, and relaxation to the ground state with photon emission. Despite their similarity, the color and intensity of the emitted bioluminescence and the dependency on pH and other molecules (e.g., ATP) differ among luciferase–luciferin pairs. Therefore, the selection of appropriate luciferase–luciferin pairs according to the biological process of interest is crucially important for BLI.
Typical natural luciferases emit weak bioluminescence with wavelengths shorter than 650 nm. Therefore, the major obstacles hindering the wider use of in vivo BLI have been the absorption and scattering of emitted bioluminescence by the host tissue. To overcome the defects, many studies have been conducted to yield brighter, red-shifted bioluminescence systems by genetic or chemical engineering. This review briefly explains recent advances in the development of engineered bioluminescence systems. To elucidate the improvements of bioluminescence intensity, we first specifically describe the mutagenic engineering on FLuc, RLuc, and OLuc. Then, we introduce another approach for enhancing bioluminescence intensity using fluorescent proteins, which is based on bioluminescence resonance energy transfer (BRET). Regarding wavelength engineering, we introduce both genetic luciferase manipulation and chemical luciferin engineering. We offer conclusions and perspectives for the future improvement of bioluminescence systems.

Figure 1. 3D structures of luciferases used in in vivo bioluminescence imaging (BLI). (a) Photinus pyralis firefly luciferase (FLuc); PDB ID: 5DVQ. (b) Renilla reniformis luciferase (RLuc); PDB ID: 2PSD. (c) NanoLuc luciferase (NLuc); PDB ID: 5IBO.
Figure 2. Chemical structures of d-luciferin and its red-shifted derivatives: (a) d-luciferin, (b) NH2-NpLH2, (c) OH-NpLH2, (d) 6′-aminoluciferin, (e) CycLuc1, (f) CycLuc2, (g) CycLuc7, (h) CycLuc10 (i) Infra-luciferin, (j) Akalumine-HCl, (k) PhOH-Luc.
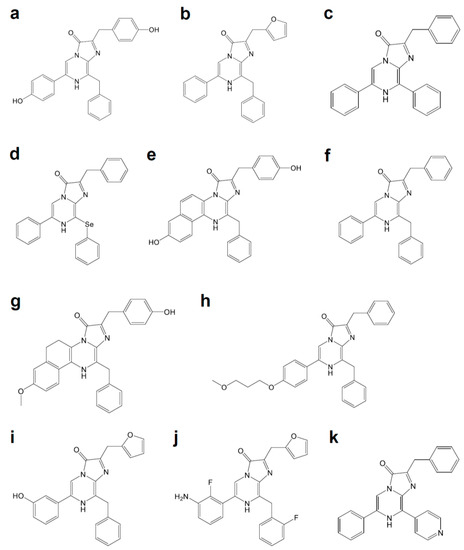
Figure 3. Coelenterazine derivatives for brighter or red-shifted bioluminescence systems: (a) Coelenterazine, (b) Furimazine, (c) Diphenylterazine, (d) Selenoterazine, (e) Coelenterazine-v, (f) DeepBlueC, (g) methoxy-eCoelenterazine, (h) BBlue2.3, (i) Hydrofurimazine, (j) Fluorofurimazine, (k) 8pyDTZ.