N6-methyladenosine (m6A) is the most prevalent modification in the mRNAs of many eukaryotic species. The abundance and effects of m6A are determined by dynamic interactions between its methyltransferases (“writers”), demethylases (“erasers”), and binding proteins (“readers”). It has been indicated that there is a strong correlation between m6A and virus infection in mammals. In the case of plant virus infection, it appears that m6A plays a dual role. On the one hand, m6A acts as a plant immune response induced by virus infection, inhibiting viral replication or translation through methylation of viral genome RNAs. On the other hand, m6A acts as part of an infection strategy employed by plant viruses to overcome the host immune system by interacting with m6A-related proteins.
- m6A methylation
- plant viruses
- defense mechanism
1. Plant Viruses May Act as an Inducer to Disrupt m6A Methylation
2. Plant Viral RNA Can Be the Target of m6A Methylation
Methylation of m6A was shown to be conserved in the genomic RNAs of diverse mammalian viruses [5][6][7]. In plant viruses, the presence of m6A in the genomes of two members of the Bromoviridae family, AMV and cucumbercucumber mosaic mosaic virusvirus (CMV), has been reported [4]. In the case of AMV, viral accumulation was reduced in inoculated leaves of a m6A demethylase (atALKBH9B) mutant in Arabidopsis. Whereas, higher m6A levels in AMV genomic RNAs were observed in these mutant plants. This suggests that m6A modification negatively affects viral infection. They found that the CMV genome also contains m6A but that it differs from AMV. The abundance of m6A methylation in viral RNA and virus infection were modified in atALKBH9B mutant plants, which might be due to the fact that the CMV coat protein (CP) did not interact with atALKBH9B in vivo [4]. This suggests a similar feature of m6A methylation in viruses that replicate in the cytoplasm of plant cells and mammalian. However, with these data, people know little about the status of m6A methylation in the AMV and CMV genome RNA. According to the prediction and identification of m6A methylation in viral RNA, it is mainly concentrated in the CDS region, while m6A methylation of plant endogenous gene mRNA is mostly concentrated in the 3’UTR of TMV [3]. This indicates that before the plant was successfully infected, the 5’ and 3’UTRs of the virus had been modified by the m6A methylation mechanism of the plant, resulting in unsuccessful viral replication and/or translation. In this case, m6A methylation in the CDS region has been detected in viral RNA as well, indicating that these methylations may not influence virus replication and proliferation but act as a protective mechanism for viral RNA and protect them from being degraded by RNase. The other question that remains unclear is whether the m6A methylations affect the translation of viral RNA.3. m6A Methylation Is One of the Defense Mechanisms against Plant Viral Infection
In mammals, RNA m6A methylation is catalyzed by a polyprotein complex composed of METTL3, METTL14, Wilms’ tumor 1-associating protein (WTAP), the human homolog of Drosophila Virilizer (KIAA1429) [14], and several cofactors not yet identified [15][16].
In some cases, viral infection are positively (in the case of hepatitis C virus, HCV) and negatively (in the case of Zika virus, ZIKV) regulated by knockdown of METTL3/14 and ALKBH5, or FTO (fat mass and obesity-associated protein), respectively [6][17]. Although m6A has long been known to exist in plant mRNAs, the proteins involved in m6A methylation have only recently been detected through mutant analysis, homology search, and mRNA interactome capture in Arabidopsis thaliana [6][17]. The review by Marlene Reichel et al., showed that the orthologues of several methylosome subunits have been identified and were shown to interact with each other. METHYLTRANSFERASE A (MTA), a METTL3 homolog that plays a critical role in plant development in Arabidopsis, has been identified [18][19]. The Arabidopsis FIP37 protein, a plant homolog of WTAP interacting with MTA both in vitro and in vivo, is essential for mediating m6A mRNA modification of key shoot meristem genes [18][20]. In the study of a protein with homology to VIRMA/KIAA1429 involved in m6A formation in mammals [14], m6A levels in the vir-1 mutant were reduced to approximately 10%, and the mutant showed aberrant formation of lateral roots and root caps as well as aberrant cotyledon development [21]. METHYLTRANSFERASE B (MTB), an orthologue of human METTL14 [21] may display enzymatic activity in Arabidopsis [22]. m6A levels were reduced to 50% in an inducible MTB RNAi line [21]. An additional component of the Arabidopsis writer complex was HAKAI, the orthologue of an E3 ubiquitin ligase. The m6A levels are reduced to 35% in hakai mutant lines without obvious phenotypes [21]. The Arabidopsis homolog of RBM15 is FPA, which regulates the flowering time by RNA-mediated chromatin silencing of the floral repressor FLOWERING LOCUS C (FLC) [23][24].
4. The Virus Encodes AlkB Protein to Promote Virus Infection
Escherichia coli AlkB proteins (members of the 2-oxoglutarate (2OG)- and Fe(II)-dependent oxygenase superfamily) are involved in DNA and RNA repair [30][31][32]. Eukaryotes usually have several proteins encoding ALKB-like genes. Nine ALKB homologs have been identified in mammals: ALKBH1-8 and FTO. Among them, ALKB5 is well studied. In HCV and ZIKV, viral titer was negatively affected when m6A modification of their genomic RNAs were regulated by the knockdown of ALKBH5 and FTO. m6A abundance in the ZIKV genome negatively affected the viral titer [6]. The production of infectious virus decreased when subjected by the depletion of FTO but not ALKBH5 [17]. The Arabidopsis genome contains 13 homologs (atALKBH1-10B) of E. coli AlkB [33]. According to the subcellular localization assay, all these proteins display a nucleocytoplasmic localization pattern except for atALKBH1D, which localizes to the chloroplast as well, and atALKBH9B, which is exclusively cytoplasmic [33]. The function of most proteins remains unknown. It has been demonstrated that the demethylase activity of atALKBH9B modulates viral infection of AMV but not of CMV. Whereas, it appears that atALKBH10B is involved in the regulatory network of floral transition in Arabidopsis [34][35]. This indicates that the host RNA methyltransferase machinery may represent an additional host regulatory mechanism to counter infection by viruses. Several plant viruses have been found to contain ALKB protein homologs or domains, suggesting a counter-defense mechanism exerted by these viruses. In 2005, plant virologists found ALKB-like domains in 22 different single-stranded RNA positive-stranded plant viruses based on protein library sequence alignments.| Virus. | M6A-Related Proteins | Summary of Knowledge | References |
|---|---|---|---|
| Aalfalfa mosaic virus (AMV) | ALKBH9B (At2g17970) | The demethylation activity of atALKBH9B affected the infectivity of AMV by interacting with CP of AMV. Suppression of atALKBH9B increased the relative abundance of m6A in the AMVgenome, impairing the systemic invasion of the plant. | Martinez-Perez et al., 2017 |
| Cucumber mosaic virus (CMV) | ALKBH9B (At2g17970) | atALKBH9B does not have any effect on CMV infection. atALKBH9B does not interact with CP of CMV. | Martinez-Perez et al., 2017 |
| Tobacco mosaic virus (TMV) | The potential demethylase XM_009801708 in Nicotiana tabacum. | The overall level of m6A decreases after (TMV) infection in Nicotiana tabacum. The expression level of XM_009801708 is increased upon TMV infection. |
Zhurui et al., 2018 |
| Grapevine virus A (GVA) | Containing ALKB domain in viral genome | Maintaining the integrity of the viral RNA genome through removal of deleterious RNA damage. | Van den Born et al., 2008 |
| Blueberry scorch virus (BlScV) | Containing ALKB domain in viral genome | Maintaining the integrity of the viral RNA genome through removal of deleterious RNA damage. | Van den Born et al., 2008 |
| Blackberry virusY (BVY) | Containing ALKB domain in viral genome | Maintaining the integrity of the viral RNA genome through removal of deleterious RNA damage. | Van den Born et al., 2008 |
| Little cherry virus (LChV-2) | Containing ALKB domain in viral genome | Not tested. | Van den Born et al., 2008 |
| Citric leave blotch virus (CLBV) | Containing ALKB domain in viral genome | Not tested. | Van den Born et al., 2008 |
| Chrysanthemum virus B (CVB) | Containing ALKB domain in viral genome | Not tested. | Van den Born et al., 2008 |
| Lily symptomless virus (LSV) | Containing ALKB domain in viral genome | Not tested. | Van den Born et al., 2008 |
| Apple stem pitting virus (ASPV) | Containing ALKB domain in viral genome | Not tested. | Van den Born et al., 2008 |
| Garlic latent virus (GLV) | Containing ALKB domain in viral genome | Not tested. | Van den Born et al., 2008 |
| Zygocactus virusX (ZVX) | Containing ALKB domain in viral genome | Not tested. | Van den Born et al., 2008 |
| Burdock mottle virus (BdMoV) | Containing ALKB domain in viral genome | Not tested. | Kondo et al., 2013 |
| Black raspberry necrosis virus (BRNV) | Containing ALKB domain in viral genome | Not tested. | McGavin et al., 2010 |
5. The Interplay between m6A Methylation and Viruses in Plant
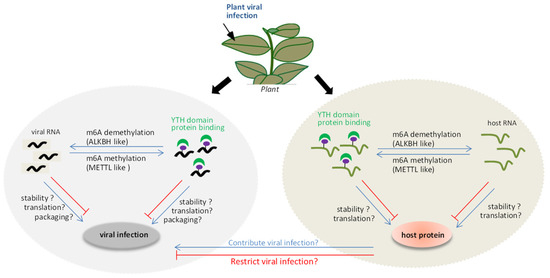
A model of interplay between the m6A methylation and plant viruses (Figure 1) was proposed. On the one hand, the m6A methylation system can be used as a defense mechanism for the host to resist foreign invasion upon viral invasion. The viral RNA could be methylated by METTL-like proteins, together with YTH domain proteins, affecting the stability, translation, or viral particle packaging during viral infection. Alternatively, the positions of m6A methylation in the viral RNA directly inhibit the replication conducted by virus replicase, which affects viral replication. On the other hand, viral RNA may also induce an imbalance in the m6A methylation and demethylation status of the host endogenous genes to regulate its expression. This can, in turn, indirectly influence the viral infection. However, several mysteries remain to be clarified, such as (i) how plants utilize the m6A methylation mechanism to modulate viral infection paying special attention to the relevance of proteins associated with m6A methylation, such as ALKBH5-like, METTL21A-like, and YTH domain proteins, to viral RNA stability and infection; (ii) how plant viruses act as inducers of responses that disrupt the reversible balance of the m6A methylation system, resulting in m6A methylation changes in host genes during plant viral infection.
References
- Liu, J.; Xu, Y.P.; Li, K.; Ye, Q.; Zhou, H.Y.; Sun, H.; Li, X.; Yu, L.; Deng, Y.Q.; Li, R.T. The methylome of SARS-CoV-2 in host cells. Cell Res. 2021, 31, 404–414.
- Tirumuru, N.; Wu, L. HIV-1 envelope proteins up-regulate N6 -methyladenosine levels of cellular RNA independently of viral replication. J. Biol. Chem. 2019, 294, 3249–3260.
- Li, Z.; Shi, J.; Lu, Y.; Zhao, X.; Ran, L.; Hu, D.; Song, B. N6 -methyl-adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. Virol. J. 2018, 15, 87.
- Martínez-Pérez, M.; Aparicio, F.; MP, L.; Bellés, J.M.; JA, S.; Pallás, V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760.
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 16011.
- Lichinchi, G.; Zhao, B.S.; Wu, Y.; Lu, Z.; Qin, Y.; He, C.; Rana, T. Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe 2016, 20, 666–673.
- Kennedy, E.M.; Bogerd, H.P.; Kornepati, A.V.R.; Kang, D.; Ghoshal, D.; Marshall, J.B.; Poling, B.C.; Tsai, K.; Gokhale, N.S.; Horner, S.M. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 2016, 19, 675–685.
- Tan, B.; Gao, S.J. The RNA Epitranscriptome of DNA Viruses. J. Virol. 2018, 92, e00696-18.
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export -NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017, 27, 606–625.
- Canaani, D.; Kahana, C.; Lavi, S.; Groner, Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 1979, 6, 2879–2899.
- Aloni, Y.; Dhar, R.; Khoury, G. Methylation of nuclear simian virus 40 RNAs. J. Virol. 1979, 32, 52–60.
- Finkel, D.; Groner, Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology 1983, 131, 409–425.
- Hesser, C.R.; Karijolich, J.; Dominissini, D.; He, C.; Glaunsinger, B.A. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 2018, 14, e1006995.
- Schwartz, S.; Mumbach, M.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D. Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5’Sites. Cell Rep. 2014, 8, 284–296.
- Duan, H.C.; Wang, Y.; Jia, G. Dynamic and reversible RNA N6-methyladenosine methylation. Wiley Interdiscip. Rev. RNA 2019, 10, e1507.
- Zhang, W.; Qian, Y.; Jia, G. The detection and functions of RNA modification m6A based on m6A writers and erasers. J. Biol. Chem. 2021, 297, 100973.
- Gokhale, N.; Mcintyre, A.R.; Mcfadden, M.; Roder, A.; Horner, S. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 2016, 20, 654–665.
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288.
- Bodi, Z.; Zhong, S.; Mehra, S.; Song, J.; Graham, N.; Li, H.; May, S.; Fray, R.G. Adenosine Methylation in Arabidopsis mRNA is Associated with the 3’End and Reduced Levels Cause Developmental Defects. Front. Plant 2012, 3, 48.
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cell 2016, 38, 186–200.
- Růžička, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S.; et al. Identification of factors required for m(6)A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017, 215, 157–172.
- Balacco, D.L.; Soller, M. The m(6)A Writer: Rise of a Machine for Growing Tasks. Biochemistry 2019, 58, 363–378.
- Baeurle, I.; Smith, L.; Baulcombe, D.C.; Dean, C. Widespread Role for the Flowering-Time Regulators FCA and FPA in RNA-Mediated Chromatin Silencing. Science 2007, 318, 109–112.
- Hornyik, C.; Duc, C.; Rataj, K.; Terzi, L.C.; Simpson, G.G. Alternative polyadenylation of antisense RNAs and flowering time control. Biochem. Soc. Trans. 2010, 38, 1077–1081.
- Li, Y.; Wang, X.; Li, C.; Hu, S.; Yu, J.; Song, S. Transcriptome-wide N6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 2014, 11, 1180–1188.
- Wei, L.H.; Song, P.; Wang, Y.; Lu, Z.; Tang, Q.; Yu, Q.; Xiao, Y.; Zhang, X.; Duan, H.C.; Jia, G. The m6A Reader ECT2 Controls Trichome Morphology by Affecting mRNA Stability in Arabidopsis. Plant Cell 2018, 30, 968–985.
- Arribas-Hernández, L.; Bressendorff, S.; Hansen, M.H.; Poulsen, C.; Erdmann, S.; Brodersen, P. An m6A-YTH Module Controls Developmental Timing and Morphogenesis in Arabidopsis. Plant Cell 2018, 30, 952–967.
- Zhou, J.; Ji, W.; Gao, X.; Zhang, X.; Qian, S.B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594.
- Zhang, T.Y.; Wang, Z.Q.; Hu, H.C.; Chen, Z.Q.; Liu, P.; Gao, S.Q.; Zhang, F.; He, L.; Jin, P.; Xu, M.Z.; et al. Transcriptome-Wide N(6)-Methyladenosine (m6A) Profiling of Susceptible and Resistant Wheat Varieties Reveals the Involvement of Variety-Specific m6A Modification Involved in Virus-Host Interaction Pathways. Front. Microbiol. 2021, 12, 656302.
- Aravind, L.; Koonin, E.V. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001, 2, 849–861.
- Falnes, P.; Johansen, R.F.; Seeberg, E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 2002, 419, 178–182.
- Trewick, S.C.; Henshaw, T.F.; Hausinger, R.P.; Lindahl, T.; Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 2002, 419, 178–182.
- Mielecki, D.; Zugaj, D.Ł.; Muszewska, A.; Piwowarski, J.; Chojnacka, A.; Mielecki, M.; Nieminuszczy, J.; Grynberg, M.; Grzesiuk, E. Novel AlkB dioxygenases-alternative models for in silico and in vivo studies. PLoS ONE 2012, 7, e30588.
- Duan, H.C.; Wei, L.H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B is An RNA N6-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell 2017, 29, 2995–3011.
- Reichel, M.; Kster, T.; Staiger, D. Marking RNA: m6A writers, readers and functions in Arabidopsis. J. Mol. Cell. Biol. 2019, 11, 899–910.
