The Xishuangbanna (XIS) cucumber is an important botanical variety, accumulating high levels of β-carotene (700 μg/100 g) in the endocarp of mature fruit compared with normal green/white flesh types (25–50 μg/100 g, fresh weight). β-carotene is an essential precursor of provitamin A synthesis required for human health, thus XIS cucumber is an appealing germplasm for vitamin A breeding programs. In this review, we highlighted the molecular research progress of XIS cucumber as well as the bottlenecks undermining its utilization in genetic breeding. The XIS cucumber was first reported in 1983; thereafter, the literature on XIS cucumber was sporadic until 2012 and 2013 following QTL mapping of the Ore gene and subsequent cloning of the CsaBCH1 gene, respectively. Whereas QTL mapping studies underlying its flowering time and fruit quality-related traits have been reported, fine mapping of their candidate genes remains unknown. Cucumber fruits are mainly consumed at the fresh immature stage; however, XIS cucumber accumulates β-carotene at mature fruit state, thus limiting the utilization of β-carotene derived from it. In our opinion, we believe that the production and commercialization of immature orange-fleshed cucumber would gain wider acceptance among consumers. Additionally, we highlighted a comprehensive breeding strategy, precisely for enhanced β-carotene accumulation based on prior studies of XIS cucumber coupled with those from other crops. In MAS, we proposed a schematic molecular backcross breeding strategy using lines possessing both ore and fft1.1 loci. This review, therefore, provides insights into XIS cucumber research and opportunities for further genetic breeding
The Xishuangbanna (XIS) cucumber is an important botanical variety, accumulating high levels of β-carotene (700 μg/100 g) in the endocarp of mature fruit compared with normal green/white flesh types (25–50 μg/100 g, fresh weight). β-carotene is an essential precursor of provitamin A synthesis required for human health, thus XIS cucumber is an appealing germplasm for vitamin A breeding programs.
- Xishuangbanna cucumber
- molecular breeding
- beta-carotene
- Nutritional quality
1. Introduction
2. Chronological Perspective of Orange-Fleshed XIS Cucumber Discovery and Development
3. Research Achievements Derived from the Utilization of Orange-Fleshed Cucumber and Research Gaps
3.1. Agronomic Characteristics of Orange-Fleshed Cultivar and Associated QTL
3.1.1. XIS Cucumber Vegetative Growth and Flowering Characteristics
3.1.2. XIS Cucumber Fruit Quality Characteristics

3.2. Molecular Markers and QTL Analysis Related to Flesh Color and Quantity of β-Carotene Content in Cucumber
3.2.1. General Overview of Molecular Markers
|
Variable |
Breeding Technique |
Pop. |
Chr. |
QTL |
Genes/Markers |
Predicted Functions |
Ref. |
|
|
Orange flesh |
QTL mapping |
F2 |
6 3 |
mc6.1/ec6. mc3.1/ec3.1 |
|
|
[20] |
|
|
β-carotene (Endocarp) |
QTL (mapping) (SSR markers) |
RIL-F9, |
3 |
ec3.1 |
Ore gene |
|
[23] |
|
|
Cloning |
|
3 |
ec3.1 |
CsBCH1 (CsGy3G017310.1) |
β-carotene hydroxylase |
[1] |
||
|
Yellow flesh (Mesocarp) |
QTL (mapping) SSR and indel |
F2 |
7 |
yf7.1 |
yfSSR108,yfIndel29), 21 predicted candidate genes |
|
[25] |
|
|
Round fruit shape |
QTL mapping |
F2,F2.3 |
|
FS5.2 |
|
|
[28] |
|
|
Fruit diameter |
QTL mapping |
F2 |
1 |
fd1.1 |
|
|
[28,38] |
|
|
Fruit length |
Mapping |
F2 |
3,5, 6 |
fl3.1, fl5.1, fl6.1 |
|
- |
[38] [27] [28] |
|
|
Length to diameter ratio |
QTL mapping |
F2 |
1 |
ldl1.1 |
|
|
[28] |
|
|
Fruit weight |
QTL |
RIL-F9 |
|
fw6.1, fw4.1, fw2.1 |
|
|
[27] |
|
|
Photoperiod flowering time |
QTL mapping |
F2, F2.3 |
6 |
FT6.1 |
|
|
[28] |
|
|
Male flowering time |
QTL mapping |
F2 |
1 6 |
mft1.1 mft6.1 |
|
|
[38] [28] |
|
|
Female flowering time |
QTL mapping
QTL, Transcriptomic |
F2
RIL-F9 |
1,5,6
1 |
fft1.1, fft5.1,fft6.2
dff1.1 |
CsaNFYA1 (Csa1G613580) |
Integrate multiple gene |
[27,28,38]
[29] |
|
3.2.2. Molecular Markers and QTL Analysis in XIS Cucumber
3.3. Regulation of Carotenoid Biosynthesis and Accumulation
4. Future Prospects in Molecular Breeding of β-Carotene Content in Cucumber
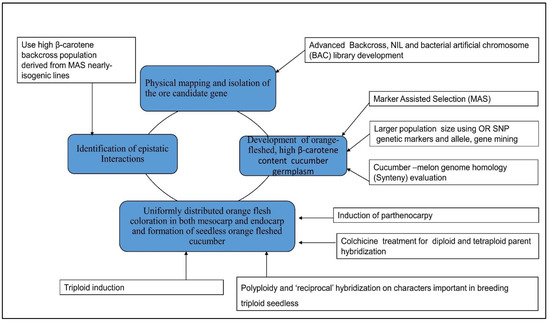
4.1. Combined Omics and Gene Editing Technology for Carotenoid Improvement
4.2. Candidate Gene Approach and Association Mapping Applications in Carotenoid Nutritional Improvement
4.3. Manipulation of Hormonal and Environmental Cues Involved in the Regulation of Carotenoid Biosynthesis
4.4. Application of Marker-Assisted Selection for β-Carotene Improvement and Development of Immature Orange-Fleshed Cucumber Variety
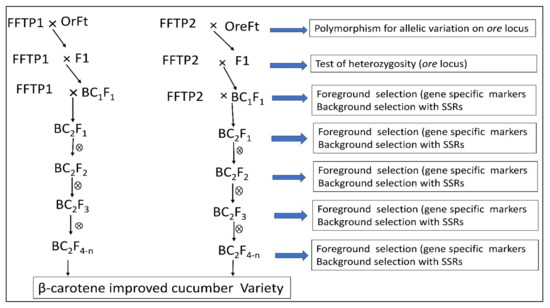
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qi, J.; Liu, X.; Shen, D.; Miao, H.; Xie, B.; Li, X.; Zeng, P.; Wang, S.; Shang, Y.; Gu, X.; et al. A genomic variation map provides insights into the genetic basis of cucumber domestication diversity. Nat. Genet. 2013, 45, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Chomicki, G.; Schaefer, H.; Renner, S.S. Origin domestication of Cucurbitaceae crops: Insights from phylogenies, genomics archaeology. New Phytol. 2020, 22, 1240–1255. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y. Cucumis sativus Chromosome Evolution, Domestication, Genetic Diversity: Implications for Cucumber Breeding. Plant Breed. Rev. 2021, 44, 79–111. [Google Scholar] [CrossRef]
- Lv, J.; Qi, J.; Shi, Q.; Shen, D.; Zhang, S.; Shao, G.; Li, H.; Sun, Z.; Weng, Y.; Shang, Y.; et al. Genetic diversity population structure of cucumber (Cucumis sativus L.). PLoS ONE 2012, 7, e46919. [Google Scholar] [CrossRef]
- Sharma, S.; Kumar, R.; Sharma, H.R.; Sharma, A.; Gautam, N. Divergence Studies for Different Horticultural Traits in Cucumber (Cucumis sativus L.). Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1733–1741. [Google Scholar] [CrossRef]
- Priyadarshini, M.; Das, S.; Muduli, K.C.; Mohanty, S.; Sahoo, S.; Pradhan, B.R. Characterization of cucumber genotypes through seed morphological characters. J. Pharm. Phytochem. 2012, 10, 2158–2161. [Google Scholar]
- Butnariu, M.; Butu, A. Chemical composition of Vegetables their products. Handb. Food Chem. 2015, 20, 627–692. [Google Scholar]
- Aderinola, T.A.; Abaire, K.E. Quality acceptability, nutritional composition antioxidant properties of carrot-cucumber juice. Beverages 2019, 5, 15. [Google Scholar] [CrossRef]
- Available online: https://www.haifa-group.com/files/Guides/Cucumber.pdf (accessed on 11 January 2022).
- Chen, J.F.; Zhang, S.L.; Zhang, X.G. The Xishuangbanna gourd (Cucumis sativus var. xishuangbannesis Qi et Yuan), a traditionally cultivated plant of the Hanai People, Xishuangbanna, Yunnan, China. Cucurbit Genet. Coop. Rpt. 1994, 17, 18–20. [Google Scholar]
- Navazio, J.P.; Simon, P.W. Diallel analysis of high carotenoid content in cucumbers. J. Am. Soc. Hortic. Sci. 2001, 126, 100–104. [Google Scholar] [CrossRef]
- Ranjan, P.; Pey, A.; Munshi, A.D.; Bhardwaj, R.; Gangopadhyay, K.K.; Malav, P.K.; Kumar, A. Orange-fleshed cucumber (Cucumis sativus var. sativus L.) germplasm from North-East India: Agro-morphological, biochemical evolutionary studies. Genet. Resour. Crop Evol. 2019, 66, 1217–1230. [Google Scholar] [CrossRef]
- Renner, S.S. A valid name for the Xishuangbanna gourd, a cucumber with carotene-rich fruits. PhytoKeys 2017, 85, 87. [Google Scholar] [CrossRef]
- Burger, Y.; Paris, H.S.; Cohen, R.; Katzir, N.; Tadmor, Y.; Lewinsohn, E.; Schaffer, A.A. 3 Genetic Diversity of Cucumis melo. Hort. Rev. 2010, 36, 165–198. [Google Scholar]
- Wiseman, E.M.; Bar-El Dadon, S.; Reifen, R. The vicious cycle of vitamin a deficiency: A review. Crit. Rev. Food Sci. Nutri. 2017, 57, 3703–3714. [Google Scholar] [CrossRef]
- Simon, P.W.; Navazio, J.P. Early orange mass 400, early orange mass 402, late orange mass 404: High β-carotene cucumber germplasm. Hort. Sci. 1997, 32, 144–145. [Google Scholar] [CrossRef]
- Qi, C.Z.; Yuan, Z.Z.; Li, Y.X. A new type of cucumber-Xishuangbanna cucumber. Acta Hortic. Sin. 1983, 10, 259–264. [Google Scholar]
- Yang, S.L.; Pu, H.; Liu, P.Y. Preliminary studies on Cucumis sativus var. xishuangbannanesis. Cucurbit Genet. Coop. Rpt. 1991, 14, 29–31. [Google Scholar]
- Navazio, J.P. Utilization of High β-Carotene Cucumber Germplasm for Genetic Improvement of Nutritional Quality. Ph.D. Thesis, University of Wisconsin, Madison, WI, USA, 1994. [Google Scholar]
- Staub, J. QTL analyses of orange color carotenoid content mapping of carotenoid biosynthesis genes in cucumber (Cucumis sativus L.). In Proceedings of the IV International Symposium on Cucurbits, Changsha, China, 21–26 September 2009; Volume 871, pp. 607–614. [Google Scholar]
- Cuevas, H.E.; Song, H.; Staub, J.E.; Simon, P.W. Inheritance of β-carotene-associated flesh color in cucumber (Cucumis sativus L.) fruit. Euphytica 2010, 17, 301–311. [Google Scholar] [CrossRef]
- Staub, J.E.; Simon, P.W.; Cuevas, H.E. Usda, Ars Eom 402-10 high β-carotene cucumber. Hortic. Sci. 2011, 46, 1426–1427. [Google Scholar] [CrossRef]
- Bo, K.; Song, H.; Shen, J.; Qian, C.; Staub, J.E.; Simon, P.W.; Chen, J. Inheritance mapping of the Ore gene controlling the quantity of β-carotene in cucumber (Cucumis sativus L.) endocarp. Mol. Breed. 2012, 30, 335–344. [Google Scholar] [CrossRef]
- Che, G.; Zhang, X. Molecular basis of cucumber fruit domestication. Curr. Opin. Plant Biol. 2019, 47, 38–46. [Google Scholar] [CrossRef]
- Lu, H.W.; Miao, H.; Tian, G.L.; Wehner, T.C.; Gu, X.F.; Zhang, S.P. Molecular mapping cidate gene analysis for yellow fruit flesh in cucumber. Mol. Breed. 2015, 35, 64. [Google Scholar] [CrossRef]
- Waters, B.M.; Kim, H.; Amundsen, K. New genetic sources for orange color in cucumber (Cucumis sativus L.) fruit flesh. bioRxiv 2019, 1, 685289. [Google Scholar]
- Bo, K.; Ma, Z.; Chen, J.; Weng, Y. Molecular mapping reveals structural rearrangements quantitative trait loci underlying traits with local adaptation in semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). Theor. Appl. Genet. 2015, 128, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Qu, S.; Bo, K.; Gao, M.; Haider, K.R.; Weng, Y. QTL mapping of domestication diversifying selection related traits in round-fruited semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis). Theor. Appl. Genet. 2017, 130, 1531–1548. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Jahn, M.; Qin, X.; Obel, H.O.; Yang, F.; Li, J.; Chen, J. Genetic Transcriptomic Analysis Reveal the Molecular Basis of Photoperiod-Regulated Flowering in Xishuangbanna Cucumber (Cucumis sativus L. var. Xishuangbannesis Qi et Yuan). Genes 2021, 12, 1064. [Google Scholar] [CrossRef] [PubMed]
- Bo, K.; Chen, L.; Qian, C.; Zhang, S.; Chen, J. Short-day treatments induce flowering of Xishuangbanna cucumber. China Cucurbits Veg. 2010, 23, 1–3. [Google Scholar]
- Xu, X.; Lu, L.; Zhu, B.; Xu, Q.; Qi, X.; Chen, X. QTL mapping of cucumber fruit flesh thickness by SLAF-seq. Sci. Rep. 2015, 5, 15829. [Google Scholar] [CrossRef]
- Garrido-Cardenas, J.A.; Mesa-Valle, C.; Manzano-Agugliaro, F. Trends in plant research using molecular markers. Planta 2018, 247, 543–557. [Google Scholar] [CrossRef]
- Kage, U.; Kumar, A.; Dhokane, D.; Karre, S.; Kushalappa, A.C. Functional molecular markers for crop improvement. Crit. Rev. Biotechnol. 2016, 36, 917–930. [Google Scholar] [CrossRef]
- Ahmad, F.; Akram, A.; Farman, K.; Abbas, T.; Bibi, A.; Khalid, S.; Waseem, M. Molecular Markers Marker Assisted Plant Breeding. Current Status their Applications in Agricultural Development. J. Environ. Agric. Sci. 2017, 11, 35–50. [Google Scholar]
- Feng, S.; Zhang, J.; Mu, Z.; Wang, Y.; Wen, C.; Wu, T.; Wang, H. Recent progress on the molecular breeding of (Cucumis sativus L.) in China. Theor. Appl. Genet. 2020, 133, 1777–1790. [Google Scholar] [CrossRef]
- Yan, J.B.; Tang, H.; Huang, Y.Q.; Zheng, Y.L.; Li, J.S. Quantitative trait loci mapping epistatic analysis for grain yield and yield components using molecular markers with an elite maize hybrid. Euphytica 2006, 149, 121–131. [Google Scholar] [CrossRef]
- Hu, J.B.; Zhou, X.Y.; Li, J.W. Development of novel EST-SSR markers for cucumber (Cucumis sativus) their transferability to related species. Sci. Hortic. 2010, 125, 534–538. [Google Scholar] [CrossRef]
- Qu, S.P.; Pan, Y.P.; Weng, Y.I.Q.U.N. QTL Mapping of flowering time and fruit shape in Xishuangbana cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). In Proceedings of the Cucurbitaceae, East Lansing, MI, USA, 21–26 October 2014; Volume 2014, pp. 54–56. [Google Scholar]
- Asins, M.J. Present future of quantitative trait locus analysis in plant breeding. Plant Breed. 2002, 121, 281–291. [Google Scholar] [CrossRef]
- Bo, K.; Wei, S.; Wang, W.; Miao, H.; Dong, S.; Zhang, S.; Gu, X. QTL mapping genome-wide association study reveal two novel loci associated with green flesh color in cucumber. BMC Plant Biol. 2019, 19, 243. [Google Scholar] [CrossRef]
- Kooistra, E. Inheritance of fruit flesh skin colors in powdery mildew resistant cucumbers (Cucumis sativus L.). Euphytica 1997, 20, 521–523. [Google Scholar] [CrossRef]
- Cunningham, F.X., Jr.; Gantt, E. Genes enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Biol. 1998, 49, 557–583. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Lado, J.; Zacarías, L.; Rodrigo, M.J. Regulation of carotenoid biosynthesis during fruit development. Carotenoids Nat. 2016, 79, 161–198. [Google Scholar] [CrossRef]
- Sun, T.; Li, L. Toward the ‘golden’ era: The status in uncovering the regulatory control of carotenoid accumulation in plants. Plant Sci. 2020, 290, 110331. [Google Scholar] [CrossRef]
- Lado, J.; Zacarías, L.; Gurrea, A.; Page, A.; Stead, A.; Rodrigo, M.J. Exploring the diversity in Citrus fruit coloration to decipher the relationship between plastid ultrastructure carotenoid composition. Planta 2015, 242, 645–661. [Google Scholar] [CrossRef]
- Li, L.; Yuan, H. Chromoplast biogenesis carotenoid accumulation. Arch. Biochem. Biophys. 2013, 539, 102–109. [Google Scholar] [CrossRef]
- Giuliano, G.; Tavazza, R.; Diretto, G.; Beyer, P.; Taylor, M.A. Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol. 2008, 26, 139–145. [Google Scholar] [CrossRef]
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 11, 3558–3563. [Google Scholar] [CrossRef]
- Yuan, H.; Owsiany, K.; Sheeja, T.; Zhou, X.; Rodriguez, C.; Li, Y.; Welsch, R.; Chayut, N.; Yang, Y.; Thannhauser, T.W.; et al. A single amino acid substitution in an ORANGE protein promotes carotenoid overaccumulation in Arabidopsis. Plant Physiol. 2015, 169, 421–431. [Google Scholar] [CrossRef]
- Lu, S.; Van Eck, J.; Zhou, X.; Lopez, A.B.; O’Halloran, D.M.; Cosman, K.M.; Li, L. The cauliflower or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 2006, 18, 3594–3605. [Google Scholar] [CrossRef]
- Tzuri, G.; Zhou, X.; Chayut, N.; Yuan, H.; Portnoy, V.; Meir, A.; Sa’Ar, U.; Baumkoler, F.; Mazourek, M.; Lewinsohn, E.; et al. A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis melo). Plant J. 2015, 82, 267–279. [Google Scholar] [CrossRef]
- Chayut, N.; Yuan, H.; Ohali, S.; Meir, A.; Sa’ar, U.; Tzuri, G.; Tadmor, Y. Distinct mechanisms of the ORANGE protein in controlling carotenoid flux. Plant Physiol. 2017, 173, 376–389. [Google Scholar] [CrossRef]
- Chayut, N.; Yuan, H.; Ohali, S.; Meir, A.; Sa’Ar, U.; Tzuri, G.; Zheng, Y.; Mazourek, M.; Gepstein, S.; Zhou, X.; et al. Syntenic relationships between cucumber (Cucumis sativus L.) melon (C. melo L.) chromosomes as revealed by comparative genetic mapping. BMC Genom. 2011, 12, 376–389. [Google Scholar] [CrossRef]
- Rajasree, V.; Pugalendhi, L. Breeding Vegetables for Nutritional Security. In Veganism—A Fashion Trend or Food as a Medicine; IntechOpen: London, UK, 2021. [Google Scholar]
- Sapir, M.; Oren-Shamir, M.; Ovadia, R.; Reuveni, M.; Evenor, D.; Tadmor, Y.; Nahon, S.; Shlomo, H.; Chen, L.; Meir, A.; et al. Molecular aspects of Anthocyanin fruit tomato in relation to high pigment-1. J. Hered. 2008, 99, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Hao, H.; Ma, L.; Zhao, C.; Yu, X. Tetraploid muskmelon alters morphological characteristics improves fruit quality. Sci. Hortic. 2010, 125, 396–400. [Google Scholar] [CrossRef]
- Singh, J.; Sharma, S.; Kaur, A.; Vikal, Y.; Cheema, A.K.; Bains, B.K.; Hossain, F. Marker-assisted pyramiding of lycopene-ε-cyclase, β-carotenehydroxylase1 opaque2 genes for development of biofortified maize hybrids. Sci. Rep. 2021, 11, 12642. [Google Scholar] [CrossRef] [PubMed]
- Robbins, M.D.; Casler, M.; Staub, J.E. Pyramiding QTL for multiple lateral branching in cucumber using nearly isogenic lines. Mol. Breed. 2008, 22, 131–139. [Google Scholar] [CrossRef]
- Staub, J.E.; Sun, Z.; Chung, S.M.; Lower, R.L. Evidence for collinearity among genetic linkage maps in cucumber. Hortic. Sci. 2007, 42, 20–27. [Google Scholar]
- Kim, J.S.; Ezura, K.; Lee, J.; Ariizumi, T.; Ezura, H. Genetic engineering of parthenocarpic tomato plants using transient SlIAA9 knockdown by novel tissue-specific promoters. Sci. Rep. 2019, 9, 18871. [Google Scholar] [CrossRef]
- Marzougui, N.; Boubaya, A.; Elfalleh, W.; Ferchichi, A.; Beji, M. Polyploidy induction in Trigonella foenum-graecum L.: Morphological chemical comparison between diploids induced autotetraploids cultivars. Acta Bot. Gall. 2009, 156, 379–389. [Google Scholar] [CrossRef]
- Mason, A.S. Polyploidy Hybridization for Crop Improvement; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef]
- Kumar, A.; Anju, T.; Kumar, S.; Chhapekar, S.; Sreedharan, S.; Singh, S.; Choi, S.; Ramchiary, N.; Lim, Y.P. Integrating Omics Gene Editing Tools for Rapid Improvement of Traditional Food Plants for Diversified Sustainable Food Security. Int. J. Mol. Sci. 2021, 22, 8093. [Google Scholar] [CrossRef]
- Brunetti, A.E.; Neto, F.C.; Vera, M.C.; Taboada, C.; Pavarini, D.P.; Bauermeister, A.; Lopes, N.P. An integrative omics perspective for the analysis of chemical signals in ecological interactions. Chem. Soc. Rev. 2018, 47, 1574–1591. [Google Scholar] [CrossRef]
- Perez-De-Castro, A.M.; Vilanova, S.; Canizares, J.; Pascual, L.; Blanca, J.; Diez, M.J.; Prohens, J.; Pico, B. Application of genomic tools in plant breeding. Curr. Genom. 2012, 13, 179–195. [Google Scholar] [CrossRef]
- Saha, S.K.; Saikot, F.K.; Rahman, M.S.; Jamal, M.A.H.M.; Rahman, S.K.; Islam, S.R.; Kim, K.H. Programmable molecular scissors: Applications of a new tool for genome editing in biotech. Mol. Ther. Nucleic Acids 2019, 14, 212–238. [Google Scholar] [CrossRef]
- Jayaraj, K.L.; Thulasidharan, N.; Antony, A.; John, M.; Augustine, R.; Chakravartty, N.; Sukumaran, S.; Maheswari, M.U.; Abraham, S.; Thomas, G.; et al. Targeted editing of tomato carotenoid isomerase reveals the role of 5′ UTR region in gene expression regulation. Plant Cell Rep. 2021, 40, 621–635. [Google Scholar] [CrossRef]
- Sun, B.; Jiang, M.; Zheng, H.; Jian, Y.; Huang, W.L.; Yuan, Q.; Tang, H.R. Color-related chlorophyll carotenoid concentrations of Chinese kale can be altered through CRISPR/Cas9 targeted editing of the carotenoid isomerase gene BoaCRTISO. Hortic. Res. 2020, 7, 161. [Google Scholar] [CrossRef]
- Thorup, T.A.; Tanyolac, B.; Livingstone, K.D.; Popovsky, S.; Paran, I.; Jahn, M. Cidate gene analysis of organ pigmentation loci in the Solanaceae. Proc. Natl. Acad. Sci. USA 2000, 97, 11192–11197. [Google Scholar] [CrossRef]
- Guzman, I.; Hamby, S.; Romero, J.; Bosl, P.W.; O’Connell, M.A. Variability of carotenoid biosynthesis in orange colored Capsicum spp. Plant Sci. 2010, 179, 49–59. [Google Scholar] [CrossRef]
- Cao, J.; Schneeberger, K.; Ossowski, S.; Günther, T.; Bender, S.; Fitz, J.; Weigel, D. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011, 43, 956–963. [Google Scholar] [CrossRef]
- Weber, A.; Clark, R.M.; Vaughn, L.; Sánchez-Gonzalez, J.D.J.; Yu, J.; Yandell, B.S.; Bradbury, P.; Doebley, J. Major regulatory genes in maize contribute to sting variation in teosinte (Zea mays ssp. parviglumis). Genetics 2011, 177, 2349–2359. [Google Scholar] [CrossRef]
- Ahmed, F.; Fanning, K.; Netzel, M.; Schenk, P.M. Induced carotenoid accumulation in Dunaliella salina Tetraselmis suecica by plant hormones UV-C radiation. Appl. Microbiol. Biotechnol. 2015, 99, 9407–9416. [Google Scholar] [CrossRef]
- Stanley, L.; Yuan, Y.W. Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef]
- Pizarro, L.; Stange, C. Light-dependent regulation of carotenoid biosynthesis in plants. Cienc. E Investig. Agrar. 2009, 36, 143–162. [Google Scholar] [CrossRef]
- Martel, C.; Vrebalov, J.; Tafelmeyer, P.; Giovannoni, J.J. The tomato MADS-box transcription factor Ripening Inhibitor interacts with promoters involved in numerous ripening processes in a Colorless Non-ripening dependent manner. Plant Physiol. 2011, 157, 1568–1579. [Google Scholar] [CrossRef]
- Pandolfini, T. Seedless fruit production by hormonal regulation of fruit set. Nutrients 2009, 1, 168–177. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]
References
- Qi, J.; Liu, X.; Shen, D.; Miao, H.; Xie, B.; Li, X.; Zeng, P.; Wang, S.; Shang, Y.; Gu, X.; et al. A genomic variation map provides insights into the genetic basis of cucumber domestication diversity. Nat. Genet. 2013, 45, 1510–1515.
- Chomicki, G.; Schaefer, H.; Renner, S.S. Origin domestication of Cucurbitaceae crops: Insights from phylogenies, genomics archaeology. New Phytol. 2020, 22, 1240–1255.
- Weng, Y. Cucumis sativus Chromosome Evolution, Domestication, Genetic Diversity: Implications for Cucumber Breeding. Plant Breed. Rev. 2021, 44, 79–111.
- Lv, J.; Qi, J.; Shi, Q.; Shen, D.; Zhang, S.; Shao, G.; Li, H.; Sun, Z.; Weng, Y.; Shang, Y.; et al. Genetic diversity population structure of cucumber (Cucumis sativus L.). PLoS ONE 2012, 7, e46919.
- Sharma, S.; Kumar, R.; Sharma, H.R.; Sharma, A.; Gautam, N. Divergence Studies for Different Horticultural Traits in Cucumber (Cucumis sativus L.). Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1733–1741.
- Priyadarshini, M.; Das, S.; Muduli, K.C.; Mohanty, S.; Sahoo, S.; Pradhan, B.R. Characterization of cucumber genotypes through seed morphological characters. J. Pharm. Phytochem. 2012, 10, 2158–2161.
- Butnariu, M.; Butu, A. Chemical composition of Vegetables their products. Handb. Food Chem. 2015, 20, 627–692.
- Aderinola, T.A.; Abaire, K.E. Quality acceptability, nutritional composition antioxidant properties of carrot-cucumber juice. Beverages 2019, 5, 15.
- Available online: https://www.haifa-group.com/files/Guides/Cucumber.pdf (accessed on 11 January 2022).
- Chen, J.F.; Zhang, S.L.; Zhang, X.G. The Xishuangbanna gourd (Cucumis sativus var. xishuangbannesis Qi et Yuan), a traditionally cultivated plant of the Hanai People, Xishuangbanna, Yunnan, China. Cucurbit Genet. Coop. Rpt. 1994, 17, 18–20.
- Navazio, J.P.; Simon, P.W. Diallel analysis of high carotenoid content in cucumbers. J. Am. Soc. Hortic. Sci. 2001, 126, 100–104.
- Ranjan, P.; Pey, A.; Munshi, A.D.; Bhardwaj, R.; Gangopadhyay, K.K.; Malav, P.K.; Kumar, A. Orange-fleshed cucumber (Cucumis sativus var. sativus L.) germplasm from North-East India: Agro-morphological, biochemical evolutionary studies. Genet. Resour. Crop Evol. 2019, 66, 1217–1230.
- Renner, S.S. A valid name for the Xishuangbanna gourd, a cucumber with carotene-rich fruits. PhytoKeys 2017, 85, 87.
- Burger, Y.; Paris, H.S.; Cohen, R.; Katzir, N.; Tadmor, Y.; Lewinsohn, E.; Schaffer, A.A. 3 Genetic Diversity of Cucumis melo. Hort. Rev. 2010, 36, 165–198.
- Wiseman, E.M.; Bar-El Dadon, S.; Reifen, R. The vicious cycle of vitamin a deficiency: A review. Crit. Rev. Food Sci. Nutri. 2017, 57, 3703–3714.
- Simon, P.W.; Navazio, J.P. Early orange mass 400, early orange mass 402, late orange mass 404: High β-carotene cucumber germplasm. Hort. Sci. 1997, 32, 144–145.
- Qi, C.Z.; Yuan, Z.Z.; Li, Y.X. A new type of cucumber-Xishuangbanna cucumber. Acta Hortic. Sin. 1983, 10, 259–264.
- Yang, S.L.; Pu, H.; Liu, P.Y. Preliminary studies on Cucumis sativus var. xishuangbannanesis. Cucurbit Genet. Coop. Rpt. 1991, 14, 29–31.
- Navazio, J.P. Utilization of High β-Carotene Cucumber Germplasm for Genetic Improvement of Nutritional Quality. Ph.D. Thesis, University of Wisconsin, Madison, WI, USA, 1994.
- Staub, J. QTL analyses of orange color carotenoid content mapping of carotenoid biosynthesis genes in cucumber (Cucumis sativus L.). In Proceedings of the IV International Symposium on Cucurbits, Changsha, China, 21–26 September 2009; Volume 871, pp. 607–614.
- Cuevas, H.E.; Song, H.; Staub, J.E.; Simon, P.W. Inheritance of β-carotene-associated flesh color in cucumber (Cucumis sativus L.) fruit. Euphytica 2010, 17, 301–311.
- Staub, J.E.; Simon, P.W.; Cuevas, H.E. Usda, Ars Eom 402-10 high β-carotene cucumber. Hortic. Sci. 2011, 46, 1426–1427.
- Bo, K.; Song, H.; Shen, J.; Qian, C.; Staub, J.E.; Simon, P.W.; Chen, J. Inheritance mapping of the Ore gene controlling the quantity of β-carotene in cucumber (Cucumis sativus L.) endocarp. Mol. Breed. 2012, 30, 335–344.
- Che, G.; Zhang, X. Molecular basis of cucumber fruit domestication. Curr. Opin. Plant Biol. 2019, 47, 38–46.
- Lu, H.W.; Miao, H.; Tian, G.L.; Wehner, T.C.; Gu, X.F.; Zhang, S.P. Molecular mapping cidate gene analysis for yellow fruit flesh in cucumber. Mol. Breed. 2015, 35, 64.
- Waters, B.M.; Kim, H.; Amundsen, K. New genetic sources for orange color in cucumber (Cucumis sativus L.) fruit flesh. bioRxiv 2019, 1, 685289.
- Bo, K.; Ma, Z.; Chen, J.; Weng, Y. Molecular mapping reveals structural rearrangements quantitative trait loci underlying traits with local adaptation in semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). Theor. Appl. Genet. 2015, 128, 25–39.
- Pan, Y.; Qu, S.; Bo, K.; Gao, M.; Haider, K.R.; Weng, Y. QTL mapping of domestication diversifying selection related traits in round-fruited semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis). Theor. Appl. Genet. 2017, 130, 1531–1548.
- Tian, Z.; Jahn, M.; Qin, X.; Obel, H.O.; Yang, F.; Li, J.; Chen, J. Genetic Transcriptomic Analysis Reveal the Molecular Basis of Photoperiod-Regulated Flowering in Xishuangbanna Cucumber (Cucumis sativus L. var. Xishuangbannesis Qi et Yuan). Genes 2021, 12, 1064.
- Bo, K.; Chen, L.; Qian, C.; Zhang, S.; Chen, J. Short-day treatments induce flowering of Xishuangbanna cucumber. China Cucurbits Veg. 2010, 23, 1–3.
- Xu, X.; Lu, L.; Zhu, B.; Xu, Q.; Qi, X.; Chen, X. QTL mapping of cucumber fruit flesh thickness by SLAF-seq. Sci. Rep. 2015, 5, 15829.
- Garrido-Cardenas, J.A.; Mesa-Valle, C.; Manzano-Agugliaro, F. Trends in plant research using molecular markers. Planta 2018, 247, 543–557.
- Kage, U.; Kumar, A.; Dhokane, D.; Karre, S.; Kushalappa, A.C. Functional molecular markers for crop improvement. Crit. Rev. Biotechnol. 2016, 36, 917–930.
- Ahmad, F.; Akram, A.; Farman, K.; Abbas, T.; Bibi, A.; Khalid, S.; Waseem, M. Molecular Markers Marker Assisted Plant Breeding. Current Status their Applications in Agricultural Development. J. Environ. Agric. Sci. 2017, 11, 35–50.
- Feng, S.; Zhang, J.; Mu, Z.; Wang, Y.; Wen, C.; Wu, T.; Wang, H. Recent progress on the molecular breeding of (Cucumis sativus L.) in China. Theor. Appl. Genet. 2020, 133, 1777–1790.
- Yan, J.B.; Tang, H.; Huang, Y.Q.; Zheng, Y.L.; Li, J.S. Quantitative trait loci mapping epistatic analysis for grain yield and yield components using molecular markers with an elite maize hybrid. Euphytica 2006, 149, 121–131.
- Hu, J.B.; Zhou, X.Y.; Li, J.W. Development of novel EST-SSR markers for cucumber (Cucumis sativus) their transferability to related species. Sci. Hortic. 2010, 125, 534–538.
- Asins, M.J. Present future of quantitative trait locus analysis in plant breeding. Plant Breed. 2002, 121, 281–291.
- Bo, K.; Wei, S.; Wang, W.; Miao, H.; Dong, S.; Zhang, S.; Gu, X. QTL mapping genome-wide association study reveal two novel loci associated with green flesh color in cucumber. BMC Plant Biol. 2019, 19, 243.
- Kooistra, E. Inheritance of fruit flesh skin colors in powdery mildew resistant cucumbers (Cucumis sativus L.). Euphytica 1997, 20, 521–523.
- Cunningham, F.X., Jr.; Gantt, E. Genes enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Biol. 1998, 49, 557–583.
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274.
- Lado, J.; Zacarías, L.; Rodrigo, M.J. Regulation of carotenoid biosynthesis during fruit development. Carotenoids Nat. 2016, 79, 161–198.
- Sun, T.; Li, L. Toward the ‘golden’ era: The status in uncovering the regulatory control of carotenoid accumulation in plants. Plant Sci. 2020, 290, 110331.
- Lado, J.; Zacarías, L.; Gurrea, A.; Page, A.; Stead, A.; Rodrigo, M.J. Exploring the diversity in Citrus fruit coloration to decipher the relationship between plastid ultrastructure carotenoid composition. Planta 2015, 242, 645–661.
- Li, L.; Yuan, H. Chromoplast biogenesis carotenoid accumulation. Arch. Biochem. Biophys. 2013, 539, 102–109.
- Giuliano, G.; Tavazza, R.; Diretto, G.; Beyer, P.; Taylor, M.A. Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol. 2008, 26, 139–145.
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 11, 3558–3563.
- Yuan, H.; Owsiany, K.; Sheeja, T.; Zhou, X.; Rodriguez, C.; Li, Y.; Welsch, R.; Chayut, N.; Yang, Y.; Thannhauser, T.W.; et al. A single amino acid substitution in an ORANGE protein promotes carotenoid overaccumulation in Arabidopsis. Plant Physiol. 2015, 169, 421–431.
- Lu, S.; Van Eck, J.; Zhou, X.; Lopez, A.B.; O’Halloran, D.M.; Cosman, K.M.; Li, L. The cauliflower or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 2006, 18, 3594–3605.
- Tzuri, G.; Zhou, X.; Chayut, N.; Yuan, H.; Portnoy, V.; Meir, A.; Sa’Ar, U.; Baumkoler, F.; Mazourek, M.; Lewinsohn, E.; et al. A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis melo). Plant J. 2015, 82, 267–279.
- Chayut, N.; Yuan, H.; Ohali, S.; Meir, A.; Sa’ar, U.; Tzuri, G.; Tadmor, Y. Distinct mechanisms of the ORANGE protein in controlling carotenoid flux. Plant Physiol. 2017, 173, 376–389.
- Chayut, N.; Yuan, H.; Ohali, S.; Meir, A.; Sa’Ar, U.; Tzuri, G.; Zheng, Y.; Mazourek, M.; Gepstein, S.; Zhou, X.; et al. Syntenic relationships between cucumber (Cucumis sativus L.) melon (C. melo L.) chromosomes as revealed by comparative genetic mapping. BMC Genom. 2011, 12, 376–389.
- Rajasree, V.; Pugalendhi, L. Breeding Vegetables for Nutritional Security. In Veganism—A Fashion Trend or Food as a Medicine; IntechOpen: London, UK, 2021.
- Sapir, M.; Oren-Shamir, M.; Ovadia, R.; Reuveni, M.; Evenor, D.; Tadmor, Y.; Nahon, S.; Shlomo, H.; Chen, L.; Meir, A.; et al. Molecular aspects of Anthocyanin fruit tomato in relation to high pigment-1. J. Hered. 2008, 99, 292–303.
- Zhang, W.; Hao, H.; Ma, L.; Zhao, C.; Yu, X. Tetraploid muskmelon alters morphological characteristics improves fruit quality. Sci. Hortic. 2010, 125, 396–400.
- Singh, J.; Sharma, S.; Kaur, A.; Vikal, Y.; Cheema, A.K.; Bains, B.K.; Hossain, F. Marker-assisted pyramiding of lycopene-ε-cyclase, β-carotenehydroxylase1 opaque2 genes for development of biofortified maize hybrids. Sci. Rep. 2021, 11, 12642.
- Robbins, M.D.; Casler, M.; Staub, J.E. Pyramiding QTL for multiple lateral branching in cucumber using nearly isogenic lines. Mol. Breed. 2008, 22, 131–139.
- Staub, J.E.; Sun, Z.; Chung, S.M.; Lower, R.L. Evidence for collinearity among genetic linkage maps in cucumber. Hortic. Sci. 2007, 42, 20–27.
- Kim, J.S.; Ezura, K.; Lee, J.; Ariizumi, T.; Ezura, H. Genetic engineering of parthenocarpic tomato plants using transient SlIAA9 knockdown by novel tissue-specific promoters. Sci. Rep. 2019, 9, 18871.
- Marzougui, N.; Boubaya, A.; Elfalleh, W.; Ferchichi, A.; Beji, M. Polyploidy induction in Trigonella foenum-graecum L.: Morphological chemical comparison between diploids induced autotetraploids cultivars. Acta Bot. Gall. 2009, 156, 379–389.
- Mason, A.S. Polyploidy Hybridization for Crop Improvement; CRC Press: Boca Raton, FL, USA, 2017.
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281.
- Kumar, A.; Anju, T.; Kumar, S.; Chhapekar, S.; Sreedharan, S.; Singh, S.; Choi, S.; Ramchiary, N.; Lim, Y.P. Integrating Omics Gene Editing Tools for Rapid Improvement of Traditional Food Plants for Diversified Sustainable Food Security. Int. J. Mol. Sci. 2021, 22, 8093.
- Brunetti, A.E.; Neto, F.C.; Vera, M.C.; Taboada, C.; Pavarini, D.P.; Bauermeister, A.; Lopes, N.P. An integrative omics perspective for the analysis of chemical signals in ecological interactions. Chem. Soc. Rev. 2018, 47, 1574–1591.
- Perez-De-Castro, A.M.; Vilanova, S.; Canizares, J.; Pascual, L.; Blanca, J.; Diez, M.J.; Prohens, J.; Pico, B. Application of genomic tools in plant breeding. Curr. Genom. 2012, 13, 179–195.
- Saha, S.K.; Saikot, F.K.; Rahman, M.S.; Jamal, M.A.H.M.; Rahman, S.K.; Islam, S.R.; Kim, K.H. Programmable molecular scissors: Applications of a new tool for genome editing in biotech. Mol. Ther. Nucleic Acids 2019, 14, 212–238.
- Jayaraj, K.L.; Thulasidharan, N.; Antony, A.; John, M.; Augustine, R.; Chakravartty, N.; Sukumaran, S.; Maheswari, M.U.; Abraham, S.; Thomas, G.; et al. Targeted editing of tomato carotenoid isomerase reveals the role of 5′ UTR region in gene expression regulation. Plant Cell Rep. 2021, 40, 621–635.
- Sun, B.; Jiang, M.; Zheng, H.; Jian, Y.; Huang, W.L.; Yuan, Q.; Tang, H.R. Color-related chlorophyll carotenoid concentrations of Chinese kale can be altered through CRISPR/Cas9 targeted editing of the carotenoid isomerase gene BoaCRTISO. Hortic. Res. 2020, 7, 161.
- Thorup, T.A.; Tanyolac, B.; Livingstone, K.D.; Popovsky, S.; Paran, I.; Jahn, M. Cidate gene analysis of organ pigmentation loci in the Solanaceae. Proc. Natl. Acad. Sci. USA 2000, 97, 11192–11197.
- Guzman, I.; Hamby, S.; Romero, J.; Bosl, P.W.; O’Connell, M.A. Variability of carotenoid biosynthesis in orange colored Capsicum spp. Plant Sci. 2010, 179, 49–59.
- Cao, J.; Schneeberger, K.; Ossowski, S.; Günther, T.; Bender, S.; Fitz, J.; Weigel, D. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011, 43, 956–963.
- Weber, A.; Clark, R.M.; Vaughn, L.; Sánchez-Gonzalez, J.D.J.; Yu, J.; Yandell, B.S.; Bradbury, P.; Doebley, J. Major regulatory genes in maize contribute to sting variation in teosinte (Zea mays ssp. parviglumis). Genetics 2011, 177, 2349–2359.
- Ahmed, F.; Fanning, K.; Netzel, M.; Schenk, P.M. Induced carotenoid accumulation in Dunaliella salina Tetraselmis suecica by plant hormones UV-C radiation. Appl. Microbiol. Biotechnol. 2015, 99, 9407–9416.
- Stanley, L.; Yuan, Y.W. Transcriptional regulation of carotenoid biosynthesis in plants: So many regulators, so little consensus. Front. Plant Sci. 2019, 10, 1017.
- Pizarro, L.; Stange, C. Light-dependent regulation of carotenoid biosynthesis in plants. Cienc. E Investig. Agrar. 2009, 36, 143–162.
- Martel, C.; Vrebalov, J.; Tafelmeyer, P.; Giovannoni, J.J. The tomato MADS-box transcription factor Ripening Inhibitor interacts with promoters involved in numerous ripening processes in a Colorless Non-ripening dependent manner. Plant Physiol. 2011, 157, 1568–1579.
- Pandolfini, T. Seedless fruit production by hormonal regulation of fruit set. Nutrients 2009, 1, 168–177.
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128.
