Various vaccinia virus (VACV) strains were applied during the smallpox vaccination campaign to eradicate the variola virus worldwide. After the eradication of smallpox, VACV gained popularity as a viral vector thanks to increasing innovations in genetic engineering and vaccine technology. Some VACV strains have been extensively used to develop vaccine candidates against various diseases. Modified vaccinia virus Ankara (MVA) is a VACV vaccine strain that offers several advantages for the development of recombinant vaccine candidates. In addition to various host-restriction genes, MVA lacks several immunomodulatory genes of which some have proven to be quite efficient in skewing the immune response in an unfavorable way to control infection in the host. Studies to manipulate these genes aim to optimize the immunogenicity and safety of MVA-based viral vector vaccine candidates.
- poxvirus
- vaccinia virus
- VACV
- viral vector vaccines
- recombinant MVA
- cross-protection
- genetic engineering
- immune-evasive genes
- host-range-related genes
1. Introduction
2. The Origin and History of Smallpox and First Smallpox Vaccination
Smallpox, caused by variola virus (VARV) major and minor [11], is an acute contagious disease that caused several hundred million deaths and devastated civilizations around the world. VARV is believed to have initially infected humans around 10,000 BCE in the early agricultural settlements of northeastern Africa [18][19] and was introduced to Europe between 400 and 600 CE [20]. Records from China in 1122 BCE and India around 1500 BCE contain descriptions that suggest smallpox or a smallpox-like disease [21] and skin lesions resembling those of smallpox were found on the mummified head of Egyptian pharaoh Ramses V [22]. In 2016, researchers isolated and reconstructed a draft genome of an antique strain of VARV from the mummy of a child who lived in Lithuania around 1650 CE. Typically, some VARV genes are fragmented and non-functional, while their homologs in VACV are sound. Comparison of these sequences to VACV showed that this elapsed VARV strain contained the same gene fragmentation pattern as modern VARV strains from the 20th century suggesting that the functional loss took place before 1650 CE [20]. More recently, VARV sequences were discovered and isolated from remains from 600 CE in Northern Europe that expose the existence of a now-extinct VARV clade [23][24]. All species of OPXV are morphologically similar and VARV major and minor are only distinguishable by real-time polymerase chain reaction (RT-PCR). Despite their similarities, they differ greatly in disease severity with case-fatality rates of 1% for variola minor and 30% for variola major [25]. While the origin of VARV and the first smallpox case in history are still uncertain, taterapox virus (TATV), which replicates in rodents, is the closest known relative of VARV. The WHO hypothesizes that rodents infected humans with an African rodent-borne variola-like or proto-variola virus about 16,000–68,000 years ago that eventually evolved into the human smallpox causing VARV [21][26]. Before vaccination was introduced in the late 1700s, variolation was used to protect individuals from smallpox. Variolation is a procedure in which healthy individuals are inoculated with powder from pustules or scabs of infected individuals who developed fewer clinical signs and symptoms in order to induce protection against smallpox infection [11]. Variolation had a significantly decreased mortality rate of 2–3%, compared to smallpox with a 15–30% mortality rate [27]. However, variolated people could transmit smallpox, particularly if the variolation site was left uncovered. This led to new outbreaks and made it clear that a better solution was sorely needed. In the second half of the 18th century, it was frequently observed that milkmaids who acquired the commonly named cowpox disease were protected against smallpox disease [11]. Similarly, Edward Jenner and others observed that the inoculation with infection material from blisters of infected cows or infected milkmaids protected humans against smallpox [28]. Although Edward Jenner may not have been the first to practice this method, he was the first to publish it [29]. Instead of pus taken from VARV lesions, he inoculated individuals with cowpox pus initially taken from a skin lesion on a milkmaid’s hand. This procedure was later called vaccination. James Phipps, the first recipient of Edward Jenner’s vaccine, did not develop any sign of disease after a challenge with smallpox and this was confirmed when sixteen other vaccinated individuals were protected against smallpox exposure. This was an important scientific landmark in the history of vaccination [30]. In the absence of any research ethics committee, Jenner used human subjects, including children, in his research [31]. There is growing evidence that Jenner was actually working with a poxvirus of equine origin that was transmitted from horses to cows instead of the cowpox virus (CPXV) [32]. In 1939, the smallpox vaccines were shown to be distinct from CPXV and were subsequently named VACV [33][34]. VACV is distinct from CPXV and VARV as it is more closely related to horsepox. However, the origin of VACV is currently unknown [13][35]. The VACV genome size is approximately 190 kbp encoding over 200 open reading frames (ORFs) [36]. The genome is bidirectional and genes are labeled R (right) and L (left) to indicate the genomic orientation of ORFs. Historically the genomes have been digested with restriction enzymes for further investigation. After digesting the genomes using HindIII the resulting 15 fragments which were obtained were specified alphabetically. Thereby, the largest fragment received the letter A and the smallest O [37][38] (Figure 1). While the historical HindIII fragment letter/ORF number nomenclature was based on the VACV strain Copenhagen [39], improved sequencing techniques and bioinformatic analysis were applied in the following years that resulted in a distinct ORF-based nomenclature for other strains such as Western Reserve (WR) [40] and MVA [41]. Based on the sequencing data, the genomes were screened for the presence of ORFs starting from the left to the right end of the genome. ORFs were numbered according to their first appearance. The first ORF found 5’ in the genome was termed 001, e.g., VACWR001 for the first ORF in the WR genome. Additionally, the ORFs were labeled R or L to indicate the genomic orientation in MVA, e.g., MVA001L for the first ORF in the genome with orientation to the left end (Figure 1 and Table 1).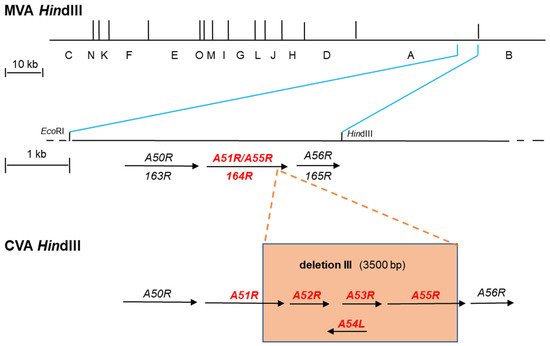
Illustration of the MVA genome and comparison of two distinct gene nomenclature systems which are currently applied. The VACV strain Copenhagen nomenclature relies on digestion of the viral genome using the restriction enzyme HindIII. In the resulting HindIII map, the fragments obtained are alphabetically labeled from A to O. The largest fragment is named A and the smallest as O. Since the genome is bidirectional, genes are labeled R (right) and L (left) to indicate the genomic orientation of ORFs. The HindIII map of the MVA genome is shown (MVA HindIII). During attenuation in cell culture, chorionallantios vaccinia virus Ankara (CVA) the parental strain of MVA, suffered amongst others six major deletions within its genome resulting in strain MVA. Deletion III is shown as an example (orange box) (CVA HindIII). This deletion is located at the right end of the viral genome. In MVA, the flanking regions of deletion III are depicted within a fragment obtained when the HindIII A fragment is cut with EcoRI. In strain Copenhagen and strain CVA this fragment harbors amongst others the genes A50R to A56R. In contrast, deletion III resulted in the loss of genes A52R to A54L, truncation of A51R, and fragmentation of A55R (orange box). The latter two genes are expressed as a fusion gene (A51R/A55R) of unknown function in MVA. Another more recent nomenclature is based on sequencing data of the full genome of MVA (177923 bp) coding sequences are numbered according to their first appearance starting from the left to the right end of the genome. The first ORF present 5’ in the genome is termed 001 and labeled R or L to indicate the genomic orientation. A more detailed illustration of the MVA genomic part after the occurrence of deletion III is shown (MVA HindIII). The genomic sequence from 146084 bp (EcoRI) to 150758 bp (HindIII) corresponds to the part obtained after cutting the HindIII A fragment with EcoRI. According to this, the nomenclature neighboring genes are hierarchically labeled including the gene 164R which corresponds to the A51R/A55R fusion gene in the Copenhagen nomenclature.
| VACV Strain | Name of Gene | Function of Gene Product | Reference |
|---|---|---|---|
| VACV COP | A40R | Type II membrane glycoprotein | [42] |
| VACV WR | A40R (VACWR165) | Acting as an immunomodulator | [18] |
| MVA | A40R (MVA152R) | Unknown function | [18] |
| VACV COP | B13R/B14R (fragmented) | - | |
| VACV WR | B13R (VACWR195) | Anti-apoptotic protein (Serine protease inhibitor 2 (SPI-2)) | [43] |
| MVA | MVA181R/182R (pseudo gene) | Non-functional protein | [43] |
| VACV COP | C6L | Inhibition of STING dimerization and phosphorylation | [44] |
| VACV WR | C6L (VACWR022) | Suppression of type I IFN—signaling pathways | [45][46] |
| MVA | C6L (MVA019L) | Inhibition of PRR signaling | [47] |
| VACV COP | C12L | Serine protease inhibitor 1 (SPI-1) | |
| VACV WR | B22R (VACWR205) | Serine protease inhibitor 1 (SPI-1) | [48] |
| MVA | Not present | - | |
| VACV COP | Not present | - | |
| VACV WR | C12L (VACWR013) | IL-18 binding protein (host defense modulator) | [49] |
| MVA | C12L (MVA008L) | IL-18 binding protein | [50] |
3. Smallpox Vaccines with Improved Safety Profile
3.1. LC16m8
LC16m8 as B5R Deletion Mutant
3.2. Dairen I Strain
3.3. Modified Vaccinia Virus Ankara (MVA)
4. Novel Viral Vector Vaccine Candidates Based on MVA with Improved Immunogenicity and Vaccine Performance
| Gene of Interest | VACV Strain and Homologous Gene |
Function of Protein | Kind of Genetic Manipulation |
Impact | References |
|---|---|---|---|---|---|
| A35R | MVA (146R) | 1. Suppression of MHC class II-restricted antigen presentation 2. Inhibition of virus-specific antibody production |
Deletion | 1. Increased production of virus-specific immunoglobulin 2. Increased numbers of virus-specific IFNγ-secreting splenocytes |
[92][93] |
| A36R | WR (VACWR159) | 1. Promotes viral egress from the host cell in association with the viral F12/E2 protein complex | Deletion | 1. Viruses lacking A36 and F12/E2 proteins form smaller plaques than mutants lacking either gene alone | [94] |
| A40R | MVA (152R) | 1. Unknown immune function | Deletion | 1. Enhanced innate immune responses in infected human macrophages 2. Increased levels of IFN-β and chemokines 3. Higher memory HIV-1-specific CD4+ and CD8+ T-cell responses |
[18] |
| A41L | WR (VACWR166) | 1. Chemokine-binding protein (vCKBP) | Deletion | 1. Enhanced virulence 2. Increased number of VACV-specific IFNγ-producing CD8+ T cells 3. More efficient cytotoxic T-cell responses in the spleen |
[95] |
| A44L | MVA (157L) | 1. 3β-HSD enzyme which catalyzes biosynthesis of steroid hormones | Deletion | 1. Simultaneous deletion with A46R and C12L (MVA008L) induced enhanced CD4+ and CD8+ T-cell responses 2. Higher levels of IL-1β, IFN-β, IFN-γ and IL-12 3. Increased functionality and proliferative capacity of memory T-cell responses |
[96] |
| A46R | MVA (159R) | 1. Inhibition of pro-inflammatory cytokine production 2. Disruption of adaptor proteins |
Deletion | 1. Simultaneous deletion with A44L and C12L (MVA008L) induced enhanced CD4+ and CD8+ T-cell responses 2. Higher levels of IL-1β, IFN-β, IFN-γ and IL-12 3. Increased functionality and proliferative capacity of memory T-cell responses |
[96] |
| B13R | MVA (181R/182R (fragmented)) WR (VACWR195) |
1. Serine protease inhibitor 2 (SPI-2) 2. Inhibition of caspase-8-mediated extrinsic apoptosis |
Insertion of functional WR gene B13R (VACWR195) into MVA |
1. Delayed apoptosis in APCs | [43] |
| C6L | MVA (019L) | 1. Blocking apoptosis by binding proapoptotic Bcl-2 proteins Bak and Bax | Deletion | 1. Deletion of C6L in rMVA-HCV induced stronger HCV NS3-specific CD8+ T-cell responses | [47] |
| C7L | WR (VACWR021) | 1. Host-range-related gene 2. Regulating viral cellular tropism |
Deletion | 1. Simultaneous deletion together with K1L gene results in abortive viral replication in murine and human cells | [97] |
| C12L | WR (VACWR205) MVA (absent) |
1. Serine protease inhibitor 1 (SPI-1) (VACWR205 corresponds to B22R, if the HindIII fragment letter/ORF number nomenclature was applied to WR) |
Deletion from WR | 1. Abortive replication in human and pig cells 2. No significant impact on replication in avian and monkey cells |
[48][98][99] |
| Insertion into MVA | 1. Increased replication in human cells | [48] | |||
| Absent instrain Copenhagen | WR (VACWR013) MVA (008L) |
1. IL-18 binding protein (VACWR013 corresponds to C12L, if the HindIII fragment letter/ORF number nomenclature was applied to WR) |
Deletionfrom MVA | 1. Simultaneous deletion with A44L and A46R genes induced enhanced CD4+ and CD8+ T-cell responses 2. Higher levels of IL-1β, IFN-β, IFN-γ and IL-12 3. Increased functionality and proliferative capacity of memory T-cell responses |
[96] |
| E3L | MVA (050L) | 1. Encoding IFN-inducible, dsRNA-activated PKR 2. Inhibiting IFN-induced activation of IFN-stimulated genes |
Deletion | 1. (MVA-ΔE3L) is unable to replicate in CEF cells | [72][73] |
| F1L | Copenhagen | 1. Inhibition of Apoptosis | Deletion | 1. Enhanced cancer cell death 2. Improved safety profile 3. No impact on the replication capacity |
[100] |
| F12L | WR (VACWR051) | 1. Promotes viral egress from host cell in association with viral A36 protein | Deletion | 1. Reduction in egress higher than by A36R deletion 2. Absence of F12 abrogates egress promoted by A36 |
[94] |
| K1L | WR (VACWR032) MVA (absent) |
1. Host-range-related gene | Deletion from WR | 1. Simultaneous deletion together with C7L gene results in abortive viral replication in murine and human cells | [97] |
| Insertion into MVA |
1. Recovery of K1 functions in MVA failed to extend host cell restriction to human cells | [101] |
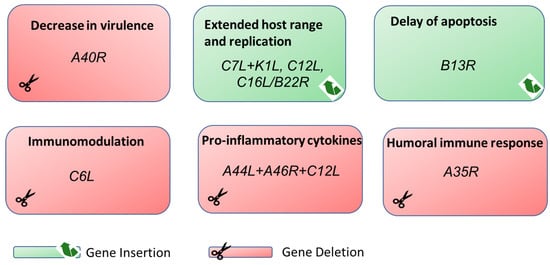
4.1. Deletion of VACV A40R Gene from MVA-B HIV-1 Vaccine Candidate Results in Increased Immunogenicity
4.2. Introduction of Host-Range-Related Genes into MVA Which Allow for Growth in Human Cells
4.3. MVA-B13R Delays Apoptosis in Antigen Presenting Cells
4.4. Removal of VACV C6L Encoding a Multifunctional Inhibitor of Type I IFN Signaling in an MVA-HCV Vaccine Candidate Elicits High Immunogenicity
4.5. Deletion of VACV A44L, A46R, and C12L Genes from the MVA Genome Improves Vector Immunogenicity and Optimizes Antigen-Specific T-Cell Responses
4.6. Deletion of A35R Increases Immunogenicity of MVA
References
- García-Arriaza, J.; Esteban, M. Enhancing Poxvirus Vectors Vaccine Immunogenicity. Hum. Vaccines Immunother. 2014, 10, 2235–2244.
- Cairns, J. The initiation of vaccinia infection. Virology 1960, 11, 603–623.
- George, C.; Katsafanas, G.C.; Moss, B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2007, 2, 221–228.
- Tolonen, N.; Doglio, L.; Schleich, S.; Krjinse Locker, J. Vaccinia Virus DNA Replication Occurs in Endoplasmic Reticulum-enclosed Cytoplasmic Mini-Nuclei. Mol. Biol. Cell. 2017, 12, 2031–2046.
- Smith, G.L.; Talbot-Cooper, C.; Lu, Y. Chapter Fourteen—How Does Vaccinia Virus Interfere with Interferon? In Advances in Virus Research; Kielian, M., Mettenleiter, T.C., Roossinck, M.J., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 100, pp. 355–378.
- Rocha, C.D.; Caetano, B.C.; Machado, A.V.; Bruna-Romero, O. Recombinant Viruses as Tools to Induce Protective Cellular Immunity against Infectious Diseases. Int. Microbiol. 2004, 7, 83–94.
- Pechenick Jowers, T.; Featherstone, R.J.; Reynolds, D.K.; Brown, H.K.; James, J.; Prescott, A.; Haga, I.R.; Beard, P.M. RAB1A Promotes Vaccinia Virus Replication by Facilitating the Production of Intracellular Enveloped Virions. Virology 2015, 475, 66–73.
- Moss, B. Smallpox Vaccines: Targets of Protective Immunity. Immunol. Rev. 2011, 239, 8–26.
- Strassburg, M.A. The Global Eradication of Smallpox. Am. J. Infect. Control 1982, 10, 53–59.
- Gilchuk, J.; Gilchuk, P.; Sapparapu, G.; Lampley, R.; Singh, V.; Kose, N.; Blum, D.L.; Hughes, L.J.; Satheshkumar, P.S.; Townsend, M.B.; et al. Cross-Neutralizing and Protective Human Antibody Specificities to Poxvirus Infections. Cell 2016, 167, 684–694.
- Fenner, F.; Henderson, D.A.; Arita, I.; Jezek, Z.; Ladnyi, I.D.; World Health Organization. Smallpox and Its Eradication; World Health Organization: Geneva, Switzerland, 1988; ISBN 978-92-4-156110-5.
- Albarnaz, J.D.; Torres, A.A.; Smith, G.L. Modulating Vaccinia Virus Immunomodulators to Improve Immunological Memory. Viruses 2018, 10, 101.
- Jacobs, B.L.; Langland, J.O.; Kibler, K.V.; Denzler, K.L.; White, S.D.; Holechek, S.A.; Wong, S.; Huynh, T.; Baskin, C.R. Vaccinia Virus Vaccines: Past, Present and Future. Antivir. Res. 2009, 84, 1–13.
- Sánchez-Sampedro, L.; Perdiguero, B.; Mejías-Pérez, E.; García-Arriaza, J.; Di Pilato, M.; Esteban, M. The Evolution of Poxvirus Vaccines. Viruses 2015, 7, 1726–1803.
- García-Arriaza, J.; Marín, M.Q.; Merchán-Rubira, J.; Mascaraque, S.M.; Medina, M.; Ávila, J.; Hernández, F.; Esteban, M. Tauopathy Analysis in P301S Mouse Model of Alzheimer Disease Immunized With DNA and MVA Poxvirus-Based Vaccines Expressing Human Full-Length 4R2N or 3RC Tau Proteins. Vaccines 2020, 8, 127.
- Smith, G.L.; Mackett, M.; Moss, B. Infectious Vaccinia Virus Recombinants That Express Hepatitis B Virus Surface Antigen. Nature 1983, 302, 490–495.
- Maki, J.; Guiot, A.-L.; Aubert, M.; Brochier, B.; Cliquet, F.; Hanlon, C.A.; King, R.; Oertli, E.H.; Rupprecht, C.E.; Schumacher, C.; et al. Oral Vaccination of Wildlife Using a Vaccinia–Rabies-Glycoprotein Recombinant Virus Vaccine (RABORAL V-RG®): A Global Review. Vet. Res. 2017, 48, 57.
- Pérez, P.; Marín, M.Q.; Lázaro-Frías, A.; Sorzano, C.Ó.S.; Gómez, C.E.; Esteban, M.; García-Arriaza, J. Deletion of Vaccinia Virus A40R Gene Improves the Immunogenicity of the HIV-1 Vaccine Candidate MVA-B. Vaccines 2020, 8, 70.
- Vieillard, V.; Combadière, B.; Tubiana, R.; Launay, O.; Pialoux, G.; Cotte, L.; Girard, P.-M.; Simon, A.; Dudoit, Y.; Reynes, J.; et al. HIV Therapeutic Vaccine Enhances Non-Exhausted CD4 + T Cells in a Randomised Phase 2 Trial. NPJ Vaccines 2019, 4, 1–9.
- Duggan, A.T.; Perdomo, M.F.; Piombino-Mascali, D.; Marciniak, S.; Poinar, D.; Emery, M.V.; Buchmann, J.P.; Duchêne, S.; Jankauskas, R.; Humphreys, M.; et al. 17th Century Variola Virus Reveals the Recent History of Smallpox. Curr. Biol. 2016, 26, 3407–3412.
- Dixon, C.W. Smallpox; J. & A. Churchhill Ltd.: London, UK, 1962.
- Hopkins, D.R. Ramses V: Earliest Know Victim? World Healthy 1980, 22–26. Available online: https://apps.who.int/iris/handle/10665/202495 (accessed on 5 July 2021).
- Spinney, L. Smallpox and Other Viruses Plagued Humans Much Earlier than Suspected. Nature 2020, 584, 30–32.
- Mühlemann, B.; Vinner, L.; Margaryan, A.; Wilhelmson, H.; de la Fuente Castro, C.; Allentoft, M.E.; de Barros Damgaard, P.; Hansen, A.J.; Nielsen, S.H.; Strand, L.M.; et al. Diverse Variola Virus (Smallpox) Strains Were Widespread in Northern Europe in the Viking Age. Science 2020, 369, eaaw8977.
- Loveless, B.M.; Mucker, E.M.; Hartmann, C.; Craw, P.D.; Huggins, J.; Kulesh, D.A. Differentiation of Variola Major and Variola Minor Variants by MGB-Eclipse Probe Melt Curves and Genotyping Analysis. Mol. Cell. Probes 2009, 23, 166–170.
- Li, Y.; Carroll, D.S.; Gardner, S.N.; Walsh, M.C.; Vitalis, E.A.; Damon, I.K. On the Origin of Smallpox: Correlating Variola Phylogenics with Historical Smallpox Records. Proc. Natl. Acad. Sci. USA 2007, 104, 15787–15792.
- Blake, J.B. Smallpox inoculation in Colonial Boston. J. Hist. Med. Allied Sci. 1953, 8, 284–300.
- Jesty, R.; Williams, G. Who invited vaccination? Malta Med. J. 2011, 23, 29–32.
- Jenner, E. An Inquiry into the Causes and Effects of the Variolae Vaccinae: A Disease Discovered in Some of the Western Counties of England, Particularly Gloucestershire, and Known by the Name of the Cow Pox; Sampson Low: London, UK, 1798.
- Baxby, D. Jenner’s Smallpox Vaccine. The Riddle of the Origin of Vaccinia Virus; Heinemann: London, UK, 1981.
- Davies, H. Ethical Reflections on Edward Jenner’s Experimental Treatment. J. Med. Ethics 2007, 33, 174–176.
- Esparza, J.; Schrick, L.; Damaso, C.R.; Nitsche, A. Equination (Inoculation of Horsepox): An Early Alternative to Vaccination (Inoculation of Cowpox) and the Potential Role of Horsepox Virus in the Origin of the Smallpox Vaccine. Vaccine 2017, 35, 7222–7230.
- Downie, A.W. The Immunological Relationship of the Virus of Spontaneous Cowpox to Vaccinia Virus. Br. J. Exp. Pathol. 1939, 20, 158–176.
- Tulman, E.R.; Delhon, G.; Afonso, C.L.; Lu, Z.; Zsak, L.; Sandybaev, N.T.; Kerembekova, U.Z.; Zaitsev, V.L.; Kutish, G.F.; Rock, D.L. Genome of Horsepox Virus. J. Virol. 2006, 80, 9244–9258.
- Qin, L.; Upton, C.; Hazes, B.; Evans, D.H. Genomic Analysis of the Vaccinia Virus Strain Variants Found in Dryvax Vaccine. J. Virol. 2011, 85, 13049–13060.
- Moss, B. Poxvirus DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010199.
- DeFilippes, F.M. Restriction Enzyme Mapping of Vaccinia Virus DNA. J. Virol. 1982, 43, 136–149.
- Mackett, M.; Archard, L.C. Conservation and Variation in Orthopoxvirus Genome Structure. J. Gen. Virol. 1979, 45, 683–701.
- Goebel, S.J.; Johnson, G.P.; Perkus, M.E.; Davis, S.W.; Winslow, J.P.; Paoletti, E. The Complete DNA Sequence of Vaccinia Virus. Virology 1990, 179, 247–266.
- Esposito, J.J.; Moss, B.; Sammons, S.A.; Frace, A.M.; Olsen-Rasmussen, M.; Osborne, J.; Baroudy, B.M.; Wohlhueter, R. Virus Pathogen Database and Analysis Resource (ViPR)—Poxviridae—Orthopoxvirus Vaccinia Virus Strain WR (Western Reserve). Available online: https://www.viprbrc.org/brc/viprStrainDetails.spg?ncbiAccession=NC_006998 (accessed on 5 July 2021).
- Antoine, G.; Scheiflinger, F.; Dorner, F.; Falkner, F.G. The Complete Genomic Sequence of the Modified Vaccinia Ankara Strain: Comparison with Other Orthopoxviruses. Virology 1998, 244, 365–396.
- Wilcock, D.; Duncan, S.A.; Traktman, P.; Zhang, W.-H.; Smith, G.L. The Vaccinia Virus A40R Gene Product Is a Nonstructural, Type II Membrane Glycoprotein That Is Expressed at the Cell Surface. J. Gen. Virol. 1999, 80 Pt 8, 2137–2148.
- Chea, L.S.; Wyatt, L.S.; Gangadhara, S.; Moss, B.; Amara, R.R. Novel Modified Vaccinia Virus Ankara Vector Expressing Anti-Apoptotic Gene B13R Delays Apoptosis and Enhances Humoral Responses. J. Virol. 2019, 93, e01648-18.
- Georgana, I.; Sumner, R.P.; Towers, G.J.; Maluquer de Motes, C. Virulent Poxviruses Inhibit DNA Sensing by Preventing STING Activation. J. Virol. 2018, 92, e02145-17.
- Unterholzner, L.; Sumner, R.P.; Baran, M.; Ren, H.; Mansur, D.S.; Bourke, N.M.; Randow, F.; Smith, G.L.; Bowie, A.G. Vaccinia Virus Protein C6 Is a Virulence Factor That Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7. PLoS Pathog. 2011, 7, e1002247.
- Stuart, J.H.; Sumner, R.P.; Lu, Y.; Snowden, J.S.; Smith, G.L. Vaccinia Virus Protein C6 Inhibits Type I IFN Signalling in the Nucleus and Binds to the Transactivation Domain of STAT2. PLoS Pathog. 2016, 12, e1005955.
- Marín, M.Q.; Pérez, P.; Gómez, C.E.; Sorzano, C.Ó.S.; Esteban, M.; García-Arriaza, J. Removal of the C6 Vaccinia Virus Interferon-β Inhibitor in the Hepatitis C Vaccine Candidate MVA-HCV Elicited in Mice High Immunogenicity in Spite of Reduced Host Gene Expression. Viruses 2018, 10, 414.
- Liu, R.; Mendez-Rios, J.D.; Peng, C.; Xiao, W.; Weisberg, A.S.; Wyatt, L.S.; Moss, B. SPI-1 Is a Missing Host-Range Factor Required for Replication of the Attenuated Modified Vaccinia Ankara (MVA) Vaccine Vector in Human Cells. PLoS Pathog. 2019, 15, e1007710.
- Reading, P.C.; Smith, G.L. Vaccinia Virus Interleukin-18-Binding Protein Promotes Virulence by Reducing Gamma Interferon Production and Natural Killer and T-Cell Activity. J. Virol. 2003, 77, 9960–9968.
- Falivene, J.; Zajac, M.P.D.M.; Pascutti, M.F.; Rodríguez, A.M.; Maeto, C.; Perdiguero, B.; Gómez, C.E.; Esteban, M.; Calamante, G.; Gherardi, M.M. Improving the MVA Vaccine Potential by Deleting the Viral Gene Coding for the IL-18 Binding Protein. PLoS ONE 2012, 7, e32220.
- Rosenthal, S.R.; Merchlinsky, M.; Kleppinger, C.; Goldenthal, K.L. Developing New Smallpox Vaccines. Emerg. Infect. Dis. 2001, 7, 920–926.
- Meseda, C.A.; Weir, J.P. Third-Generation Smallpox Vaccines: Challenges in the Absence of Clinical Smallpox. Future Microbiol. 2010, 5, 1367–1382.
- Nalca, A.; Zumbrun, E.E. ACAM2000™: The new smallpox vaccine for United States Strategic National Stockpile. Drug. Des. Devel. Ther. 2010, 4, 71–79.
- Sugimoto, M.; Yasuda, A.; Miki, K.; Morita, M.; Suzuki, K.; Uchida, N.; Hashizume, S. Gene Structures of Low-Neurovirulent Vaccinia Virus LC16m0, LC16m8, and Their Lister Original (LO) Strains. Microbiol. Immunol. 1985, 29, 421–428.
- Kenner, J.; Cameron, F.; Empig, C.; Jobes, D.V.; Gurwith, M. LC16m8: An Attenuated Smallpox Vaccine. Vaccine 2006, 24, 7009–7022.
- Saijo, M.; Ami, Y.; Suzaki, Y.; Nagata, N.; Iwata, N.; Hasegawa, H.; Ogata, M.; Fukushi, S.; Mizutani, T.; Sata, T.; et al. LC16m8, a Highly Attenuated Vaccinia Virus Vaccine Lacking Expression of the Membrane Protein B5R, Protects Monkeys from Monkeypox. J. Virol. 2006, 80, 5179–5188.
- Takahashi-Nishimaki, F.; Suzuki, K.; Morita, M.; Maruyama, T.; Miki, K.; Hashizume, S.; Sugimoto, M. Genetic Analysis of Vaccinia Virus Lister Strain and Its Attenuated Mutant LC16m8: Production of Intermediate Variants by Homologous Recombination. J. Gen. Virol. 1987, 68 Pt 10, 2705–2710.
- Smith, G.L.; Vanderplasschen, A.; Law, M. The Formation and Function of Extracellular Enveloped Vaccinia Virus. J. Gen. Virol. 2002, 83, 2915–2931.
- Bell, E.; Shamim, M.; Whitbeck, J.C.; Sfyroera, G.; Lambris, J.D.; Isaacs, S.N. Antibodies against the Extracellular Enveloped Virus B5R Protein Are Mainly Responsible for the EEV Neutralizing Capacity of Vaccinia Immune Globulin. Virology 2004, 325, 425–431.
- Empig, C.; Kenner, J.R.; Perret-Gentil, M.; Youree, B.E.; Bell, E.; Chen, A.; Gurwith, M.; Higgins, K.; Lock, M.; Rice, A.D.; et al. Highly Attenuated Smallpox Vaccine Protects Rabbits and Mice against Pathogenic Orthopoxvirus Challenge. Vaccine 2006, 24, 3686–3694.
- Ishii, K.; Ueda, Y.; Matsuo, K.; Matsuura, Y.; Kitamura, T.; Kato, K.; Izumi, Y.; Someya, K.; Ohsu, T.; Honda, M.; et al. Structural Analysis of Vaccinia Virus DIs Strain: Application as a New Replication-Deficient Viral Vector. Virology 2002, 302, 433–444.
- Mayr, A.; Munz, E. Veränderung von Vaccinevirus durch Dauerpassagen in Hühnerembryofibroblasten-Kulturen . Zentralbl. Bakteriol. Orig. 1964, 195, 24–35.
- Stickl, H.; Hochstein-Mintzel, V.; Mayr, A.; Huber, H.C.; Schäfer, H.; Holzner, A. MVA vaccination against smallpox: Clinical tests with an attenuated live vaccinia virus strain (MVA) (author’s transl). Dtsch. Med. Wochenschr. 1974, 99, 2386–2392.
- Mayr, A.; Stickl, H.; Müller, H.K.; Danner, K.; Singer, H. The smallpox vaccination strain MVA: Marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism (author’s transl). Zentralbl. Bakteriol. B 1978, 167, 375–390.
- Perkus, M.E.; Goebel, S.J.; Davis, S.W.; Johnson, G.P.; Limbach, K.; Norton, E.K.; Paoletti, E. Vaccinia Virus Host Range Genes. Virology 1990, 179, 276–286.
- Ludwig, H.; Mages, J.; Staib, C.; Lehmann, M.H.; Lang, R.; Sutter, G. Role of Viral Factor E3L in Modified Vaccinia Virus Ankara Infection of Human HeLa Cells: Regulation of the Virus Life Cycle and Identification of Differentially Expressed Host Genes. J. Virol. 2005, 79, 2584–2596.
- Beattie, E.; Tartaglia, J.; Paoletti, E. Vaccinia Virus-Encoded EIF-2 Alpha Homolog Abrogates the Antiviral Effect of Interferon. Virology 1991, 183, 419–422.
- Chang, H.W.; Watson, J.C.; Jacobs, B.L. The E3L Gene of Vaccinia Virus Encodes an Inhibitor of the Interferon-Induced, Double-Stranded RNA-Dependent Protein Kinase. Proc. Natl. Acad. Sci. USA 1992, 89, 4825–4829.
- Samuel, C.E. Antiviral Actions of Interferons. Clin. Microbiol. Rev. 2001, 14, 778–809.
- Park, C.; Peng, C.; Rahman, M.J.; Haller, S.L.; Tazi, L.; Brennan, G.; Rothenburg, S. Orthopoxvirus K3 Orthologs Show Virus- and Host-Specific Inhibition of the Antiviral Protein Kinase PKR. PLoS Pathog. 2021, 17, e1009183.
- Domingo-Gil, E.; Pérez-Jiménez, E.; Ventoso, I.; Nájera, J.L.; Esteban, M. Expression of the E3L Gene of Vaccinia Virus in Transgenic Mice Decreases Host Resistance to Vaccinia Virus and Leishmania Major Infections. J. Virol. 2008, 82, 254–267.
- Guerra, S.; Cáceres, A.; Knobeloch, K.-P.; Horak, I.; Esteban, M. Vaccinia Virus E3 Protein Prevents the Antiviral Action of ISG15. PLoS Pathog. 2008, 4, e1000096.
- Hornemann, S.; Harlin, O.; Staib, C.; Kisling, S.; Erfle, V.; Kaspers, B.; Häcker, G.; Sutter, G. Replication of Modified Vaccinia Virus Ankara in Primary Chicken Embryo Fibroblasts Requires Expression of the Interferon Resistance Gene E3L. J. Virol. 2003, 77, 8394–8407.
- Colby, C.; Duesberg, P.H. Double-Stranded RNA in Vaccinia Virus Infected Cells. Nature 1969, 222, 940–944.
- Wolferstätter, M.; Schweneker, M.; Späth, M.; Lukassen, S.; Klingenberg, M.; Brinkmann, K.; Wielert, U.; Lauterbach, H.; Hochrein, H.; Chaplin, P.; et al. Recombinant Modified Vaccinia Virus Ankara Generating Excess Early Double-Stranded RNA Transiently Activates Protein Kinase R and Triggers Enhanced Innate Immune Responses. J. Virol. 2014, 88, 14396–14411.
- Volz, A.; Sutter, G. Modified Vaccinia Virus Ankara: History, Value in Basic Research, and Current Perspectives for Vaccine Development. Adv. Virus Res. 2017, 97, 187–243.
- Drexler, I.; Staib, C.; Sutter, G. Modified Vaccinia Virus Ankara as Antigen Delivery System: How Can We Best Use Its Potential? Curr. Opin. Biotechnol. 2004, 15, 506–512.
- Drexler, I.; Heller, K.; Wahren, B.; Erfle, V.; Sutter, G. Highly Attenuated Modified Vaccinia Virus Ankara Replicates in Baby Hamster Kidney Cells, a Potential Host for Virus Propagation, but Not in Various Human Transformed and Primary Cells. J. Gen. Virol. 1998, 79 Pt 2, 347–352.
- Sutter, G.; Moss, B. Nonreplicating Vaccinia Vector Efficiently Expresses Recombinant Genes. Proc. Natl. Acad. Sci. USA 1992, 89, 10847–10851.
- Altenburg, A.F.; Kreijtz, J.H.C.M.; de Vries, R.D.; Song, F.; Fux, R.; Rimmelzwaan, G.F.; Sutter, G.; Volz, A. Modified Vaccinia Virus Ankara (MVA) as Production Platform for Vaccines against Influenza and Other Viral Respiratory Diseases. Viruses 2014, 6, 2735–2761.
- Baden, L.R.; Walsh, S.R.; Seaman, M.S.; Cohen, Y.Z.; Johnson, J.A.; Licona, J.H.; Filter, R.D.; Kleinjan, J.A.; Gothing, J.A.; Jennings, J.; et al. First-in-Human Randomized, Controlled Trial of Mosaic HIV-1 Immunogens Delivered via a Modified Vaccinia Ankara Vector. J. Infect. Dis. 2018, 218, 633–644.
- Nascimento, I.P.; Leite, L.C.C. Recombinant Vaccines and the Development of New Vaccine Strategies. Braz. J. Med. Biol. Res. 2012, 45, 1102–1111.
- Jordan, I.; Sandig, V. Matrix and Backstage: Cellular Substrates for Viral Vaccines. Viruses 2014, 6, 1672–1700.
- Panicali, D.; Paoletti, E. Construction of Poxviruses as Cloning Vectors: Insertion of the Thymidine Kinase Gene from Herpes Simplex Virus into the DNA of Infectious Vaccinia Virus. Proc. Natl. Acad. Sci. USA 1982, 79, 4927–4931.
- Mackett, M.; Smith, G.L.; Moss, B. Vaccinia Virus: A Selectable Eukaryotic Cloning and Expression Vector. Proc. Natl. Acad. Sci. USA 1982, 79, 7415–7419.
- Cottingham, M.G.; Gilbert, S.C. Rapid Generation of Markerless Recombinant MVA Vaccines by En Passant Recombineering of a Self-Excising Bacterial Artificial Chromosome. J. Virol. Methods 2010, 168, 233–236.
- Kugler, F.; Drexler, I.; Protzer, U.; Hoffmann, D.; Moeini, H. Generation of Recombinant MVA-Norovirus: A Comparison Study of Bacterial Artificial Chromosome- and Marker-Based Systems. Virol. J. 2019, 16, 100.
- Samreen, B.; Tao, S.; Tischer, K.; Adler, H.; Drexler, I. ORF6 and ORF61 Expressing MVA Vaccines Impair Early but Not Late Latency in Murine Gammaherpesvirus MHV-68 Infection. Front. Immunol. 2019, 10.
- Domi, A.; Moss, B. Engineering of a Vaccinia Virus Bacterial Artificial Chromosome in Escherichia Coli by Bacteriophage Lambda-Based Recombination. Nat. Methods 2005, 2, 95–97.
- Chiuppesi, F.; Salazar, M.d.; Contreras, H.; Nguyen, V.H.; Martinez, J.; Park, Y.; Nguyen, J.; Kha, M.; Iniguez, A.; Zhou, Q.; et al. Development of a Multi-Antigenic SARS-CoV-2 Vaccine Candidate Using a Synthetic Poxvirus Platform. Nat. Commun. 2020, 11, 6121.
- Wong, Y.C.; Croft, S.; Smith, S.A.; Lin, L.C.W.; Cukalac, T.; La Gruta, N.L.; Drexler, I.; Tscharke, D.C. Modified Vaccinia Virus Ankara Can Induce Optimal CD8+ T Cell Responses to Directly Primed Antigens Depending on Vaccine Design. J. Virol. 2019, 93, e01154-19.
- Rehm, K.E.; Roper, R.L. Deletion of the A35 Gene from Modified Vaccinia Virus Ankara Increases Immunogenicity and Isotype Switching. Vaccine 2011, 29, 3276–3283.
- Rehm, K.E.; Connor, R.F.; Jones, G.J.B.; Yimbu, K.; Roper, R.L. Vaccinia Virus A35R Inhibits MHC Class II Antigen Presentation. Virology 2010, 397, 176–186.
- Carpentier, D.C.J.; Van Loggerenberg, A.; Dieckmann, N.M.G.; Smith, G.L. Vaccinia Virus Egress Mediated by Virus Protein A36 Is Reliant on the F12 Protein. J. Gen. Virol. 2017, 98, 1500–1514.
- Clark, R.H.; Kenyon, J.C.; Bartlett, N.W.; Tscharke, D.C.; Smith, G.L. Deletion of Gene A41L Enhances Vaccinia Virus Immunogenicity and Vaccine Efficacy. J. Gen. Virol. 2006, 87, 29–38.
- Holgado, M.P.; Falivene, J.; Maeto, C.; Amigo, M.; Pascutti, M.F.; Vecchione, M.B.; Bruttomesso, A.; Calamante, G.; del Médico-Zajac, M.P.; Gherardi, M.M. Deletion of A44L, A46R and C12L Vaccinia Virus Genes from the MVA Genome Improved the Vector Immunogenicity by Modifying the Innate Immune Response Generating Enhanced and Optimized Specific T-Cell Responses. Viruses 2016, 8, 139.
- Meng, X.; Chao, J.; Xiang, Y. Identification from Diverse Mammalian Poxviruses of Host-Range Regulatory Genes Functioning Equivalently to Vaccinia Virus C7L. Virology 2008, 372, 372–383.
- Ali, A.N.; Turner, P.C.; Brooks, M.A.; Moyer, R.W. The SPI-1 Gene of Rabbitpox Virus Determines Host Range and Is Required for Hemorrhagic Pock Formation. Virology 1994, 202, 305–314.
- Kettle, S.; Blake, N.W.; Law, K.M.; Smith, G.L. Vaccinia Virus Serpins B13R (SPI-2) and B22R (SPI-1) Encode M(r) 38.5 and 40K, Intracellular Polypeptides That Do Not Affect Virus Virulence in a Murine Intranasal Model. Virology 1995, 206, 136–147.
- Pelin, A.; Foloppe, J.; Petryk, J.; Singaravelu, R.; Hussein, M.; Gossart, F.; Jennings, V.A.; Stubbert, L.J.; Foster, M.; Storbeck, C.; et al. Deletion of Apoptosis Inhibitor F1L in Vaccinia Virus Increases Safety and Oncolysis for Cancer Therapy. Mol. Ther. Oncolytics 2019, 14, 246–252.
- Wyatt, L.S.; Carroll, M.W.; Czerny, C.P.; Merchlinsky, M.; Sisler, J.R.; Moss, B. Marker Rescue of the Host Range Restriction Defects of Modified Vaccinia Virus Ankara. Virology 1998, 251, 334–342.
- Peng, C.; Moss, B. Repair of a Previously Uncharacterized Second Host-Range Gene Contributes to Full Replication of Modified Vaccinia Virus Ankara (MVA) in Human Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 3759–3767.
- Gillard, S.; Spehner, D.; Drillien, R.; Kirn, A. Localization and Sequence of a Vaccinia Virus Gene Required for Multiplication in Human Cells. Proc. Natl. Acad. Sci. USA 1986, 83, 5573–5577.
- Sivan, G.; Ormanoglu, P.; Buehler, E.C.; Martin, S.E.; Moss, B. Identification of Restriction Factors by Human Genome-Wide RNA Interference Screening of Viral Host Range Mutants Exemplified by Discovery of SAMD9 and WDR6 as Inhibitors of the Vaccinia Virus K1L-C7L- Mutant. MBio 2015, 6, e01122.
- Dobbelstein, M.; Shenk, T. Protection against Apoptosis by the Vaccinia Virus SPI-2 (B13R) Gene Product. J. Virol. 1996, 70, 6479.
- Kettle, S.; Alcamí, A.; Khanna, A.; Ehret, R.; Jassoy, C.; Smith, G.L. Vaccinia Virus Serpin B13R (SPI-2) Inhibits Interleukin-1beta-Converting Enzyme and Protects Virus-Infected Cells from TNF- and Fas-Mediated Apoptosis, but Does Not Prevent IL-1beta-Induced Fever. J. Gen. Virol. 1997, 78 Pt 3, 677–685.
- Veyer, D.L.; Maluquer de Motes, C.; Sumner, R.P.; Ludwig, L.; Johnson, B.F.; Smith, G.L. Analysis of the Anti-Apoptotic Activity of Four Vaccinia Virus Proteins Demonstrates That B13 Is the Most Potent Inhibitor in Isolation and during Viral Infection. J. Gen. Virol. 2014, 95, 2757–2768.
- Postigo, A.; Way, M. The Vaccinia Virus-Encoded Bcl-2 Homologues Do Not Act as Direct Bax Inhibitors. J. Virol. 2012, 86, 203–213.
- Lu, Y.; Stuart, J.H.; Talbot-Cooper, C.; Agrawal-Singh, S.; Huntly, B.; Smid, A.I.; Snowden, J.S.; Dupont, L.; Smith, G.L. Histone deacetylase 4 promotes type I interferon signaling, restricts DNA viruses, and is degraded via vaccinia virus protein C6. Proc. Natl. Acad. Sci. USA 2019, 116, 11997–12006.
- Frenz, T.; Waibler, Z.; Hofmann, J.; Hamdorf, M.; Lantermann, M.; Reizis, B.; Tovey, M.G.; Aichele, P.; Sutter, G.; Kalinke, U. Concomitant Type I IFN Receptor-Triggering of T Cells and of DC Is Required to Promote Maximal Modified Vaccinia Virus Ankara-Induced T-Cell Expansion. Eur. J. Immunol. 2010, 40, 2769–2777.
- Jones, C.T.; Murray, C.L.; Eastman, D.K.; Tassello, J.; Rice, C.M. Hepatitis C Virus P7 and NS2 Proteins Are Essential for Production of Infectious Virus. J. Virol. 2007, 81, 8374–8383.
- Reading, P.C.; Moore, J.B.; Smith, G.L. Steroid Hormone Synthesis by Vaccinia Virus Suppresses the Inflammatory Response to Infection. J. Exp. Med. 2003, 197, 1269–1278.
- Rasmussen, M.K.; Ekstrand, B.; Zamaratskaia, G. Regulation of 3β-Hydroxysteroid Dehydrogenase/Δ5-Δ4 Isomerase: A Review. Int. J. Mol. Sci. 2013, 14, 17926–17942.
- Perdiguero, B.; Gómez, C.E.; Pilato, M.D.; Sorzano, C.O.S.; Delaloye, J.; Roger, T.; Calandra, T.; Pantaleo, G.; Esteban, M. Deletion of the Vaccinia Virus Gene A46R, Encoding for an Inhibitor of TLR Signalling, Is an Effective Approach to Enhance the Immunogenicity in Mice of the HIV/AIDS Vaccine Candidate NYVAC-C. PLoS ONE 2013, 8, e74831.
- Nakanishi, K. Unique Action of Interleukin-18 on T Cells and Other Immune Cells. Front. Immunol. 2018, 9.
- Roper, R.L. Characterization of the Vaccinia Virus A35R Protein and Its Role in Virulence. J. Virol. 2006, 80, 306–313.
