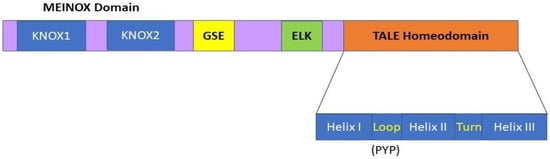
Until 2005,
KNAT7 KNAT7 was not often discussed in mutation studies of the
Class II KNOX Class II KNOX genes; however, a number of
Class II KNOX Class II KNOX mutations have recently been studied in detail (
Table 1). A T-DNA insertion in the intron of the
KNAT7 KNAT7 gene resulted in a loss-of-function mutant,
irx11 irx11, that showed only a moderately weak growth phenotype. The
irx11 irx11 mutant also exhibited the typical
irx irx phenotype in xylem vessels that were collapsed due to weak SCW formation. The
irx11 irx11 mutant did not have significantly altered cellulose or xylan content compared to controls. No lignin content of these mutants was reported at that time. While discovering a set of novel TFs involved in SCW biosynthesis, Zhong et al.
[13][12] associated
KNAT7 KNAT7 expression with SCW formation, and the dominant repression of
KNAT7 KNAT7 (DR-
KNAT7 KNAT7 mutants) affected SCW formation in both xylem and fiber cells (
Table 1). Curiously, they did not observe the typical
irx irx phenomenon in these DR-
KNAT7 KNAT7 mutants, a tell-tale sign of weak SCW formation; however, the cell wall thicknesses of both xylem vessels and fibers were reduced compared to controls (28% down in interfascicular fibers (IF), 26% down in vessels (V), and 80% down in xylary fibers (XF)). Several monosaccharides from the cell walls of DR-
KNAT7 KNAT7 mutants were reduced by 20–30%, except for arabinose, which was increased by 18%. The overexpression of
KNAT7 KNAT7 did not increase the SCW thickness of fibers and vessels. These results indicated that KNAT7 could be a positive regulator of SCW formation in
Arabidopsis Arabidopsis. However, Li et al.
[14][13] reported a contrasting observation that loss-of-function mutants in the
AtKNAT7 AtKNAT7 gene resulted in differential thicknesses of interfascicular and xylary fibers compared to vessels (58% up in IF, 35% down in V, and 31% up in XF;
Table 1). The vessels walls were thinner, resulting in collapsed xylem vessels that showed the
irx irx phenotype (similar to
[18][30]); however, the interfascicular fibers were significantly thicker than in the wild type control, suggesting that KNAT7 is a transcriptional repressor of fiber SCW formation (but a transcriptional activator of vessel SCW formation).
KNAT7 KNAT7 overexpression lines exhibited thinner fiber walls (57% down in IF) with normal vessel and xylary fiber cell walls. Interestingly, even though many SCW-specific cellulose and xylan synthesis genes were upregulated in these mutants, no quantitative changes in cellulose or xylan were reported. All ten lignin synthesis genes tested were upregulated along with an 11% increase in lignin content of cell walls from the stem. Li et al.
[18][30] speculated that KNAT7 interacts with different partner proteins in different cell types to form functionally distinct complexes. Recently, the regulatory roles of other members of the
Class II KNOX Class II KNOX gene family,
KNAT3, KNAT4, KNAT3, KNAT4, and
KNAT5 KNAT5, in SCW formation were explored in
Arabidopsis Arabidopsis inflorescence stems
[16][17][15,16] (
Table 1). Loss-of-function mutants of
knat3, knat4, knat3, knat4, and
knat5 knat5 did not produce any
irx irx phenotype, as observed in the case of loss-of-function mutants of
knat7 knat7 [16][15]. This could be due to the functional redundancy of
KNOX II KNOX II genes. However,
knat3/knat7 knat3/knat7 double mutants displayed an enhanced
irx irx phenotype compared to single
knat7 knat7 mutants. These double mutants had thinner interfascicular fiber cell walls compared to the single mutants and wild-type plants (40% down in IF) indicating a potentially positive regulatory role of KNAT3 in combination with KNAT7 in xylem SCW development. Even though many SCW genes were highly expressed in the
knat3/knat7 knat3/knat7 double mutants, the cellulose and xylan contents of their cell walls were reduced by 19% and 43%, respectively, and the changes in lignin content were not significant. The Syringyl to Guaicyl (S/G) lignin ratio was down by 83%; however, it was not possible to correlate all these cell wall content changes with the changes in gene expression patterns. In addition, the severe
irx irx phenotype in these double mutants indicated the overlapping roles and partial functional redundancy of KNAT3 and KNAT7 in xylem vessel development during SCW formation. Furthermore,
KNAT3 KNAT3 overexpression in
Arabidopsis Arabidopsis resulted in thickened interfascicular fibers in the SCW of inflorescence stems
[16][15]. This
enst
rudy described KNAT3 as a potential transcriptional activator, working together with KNAT7 to promote SCW biosynthesis in xylem vessels. A synergistic interaction of KNAT3 and KNAT7 to regulate monolignol biosynthesis in
Arabidopsis Arabidopsis was also reported in another study
[17][16]. Most importantly, they attempted to link S-lignin formation with
KNAT3 KNAT3 and
KNAT7 KNAT7 expression; however, they could not show the direct transcriptional regulation of a key gene, ferulate 5-hydroxylase (
F5H), involved in S-lignin formation by KNAT3 or KNAT7. Similar to the earlier observation by Wang et al.
[16][15], the overexpression of KNAT3 also caused thickening in the interfascicular fiber walls, indicating the positive regulation of interfascicular fiber wall development by KNAT3. These studies by Wang et al. and Qin et al.
[15,16] reconciled the paradoxical [16][17]observations about KNAT7 mutants in Arabidopsis and indicated that KNAT3 and KNAT7 might be working synergistically in fibers, but antagonistically in vessels, during the regulation of SCW biosynthesis reconciled the paradoxical observations about KNAT7 mutants in Arabidopsis and indicated that KNAT3 and KNAT7 might be working synergistically in fibers, but antagonistically in vessels, during the regulation of SCW biosynthesis ((Table 1).
Table 1.
Gene Mutations in
Class II KNOX
genes and their effect on SCW formation.
| Target Gene |
Mutation |
Type of Mutation |
Anatomy of Mutants |
References |
| AtKNAT7 |
irx11 |
T-DNA insertion |
Irregular xylem with collapsed vessels. |
[18][30] |
| AtKNAT7 |
- |
Dominant repression |
Reduced cell wall thickness of both xylem vessels and fibers; reduced composition of several monosaccharides from the cell walls. |
[13][12] |
| AtKNAT7 |
irx11 |
Loss-of-function mutation |
Thinner vessels walls resulted in a collapse of xylem vessels that showed the irx phenotype and thicker interfascicular fibers compared to controls; increase in lignin content. |
[14][13] |
| AtKNAT3, AtKNAT4, AtKNAT5 |
Single mutants |
T-DNA insertion |
No irx phenotype. |
[16][15] |
| [ | 15 | ] | [ | 14] |
KNAT3/KNAT7 |
Double mutant |
T-DNA insertion |
Enhanced irregular xylem (irx) phenotype characterized by weak inflorescence stem; reduced interfascicular fiber wall thickness and modified cell wall composition. |
[16] |
| AtKNAT7 | [ | 15 | ] |
| Arabidopsis |
Dominant repression |
Reduced expression of SCW genes that resulted in thinner fiber cell walls with altered cell wall composition. |
[ | 13][12] |
KNAT3/KNAT7 |
Double mutant |
Chimeric repression |
Thinner interfascicular fiber cell walls compared to single mutants and wild type (WT); reduced cellulose and xylan and reduced S/G lignin ratio. |
[17][16] |
| PtKNAT7 |
Poplar |
Overexpression |
OsKNAT7 |
CRISPR/CAS9 |
T-DNA insertion |
Thicker fiber cell walls; larger grain size due to cell expansion in spikelet bracts. |
[24][35] |
| GhKNL1 |
- |
Dominant repression |
Abnormal shorter fiber length. |
[22][33] |
4. Targeted Genetic Manipulations in Class II Class II KNOXKNOX Genes Confirm Their Role in SCW Formation
Apart from the detailed study of
Class II KNOX Class II KNOX gene mutants, targeted genetic manipulations of
Class II KNOX Class II KNOX genes, especially,
KNAT7 KNAT7 genes have offered some additional clues regarding the functions of these genes (
Table 2). While the overexpression of
KNAT7 KNAT7 in
Arabidopsis Arabidopsis did not produce any specific SCW phenotype
[13][12], subsequently, Li et al.
[14][13] reported that such experiments produced thin interfascicular fibers without any changes in wall thickness of vessels suggesting that KNAT7 TF is indeed a regulator of SCW formation.
Table 2.
Genetic manipulation of
Class II KNOX
genes in different plant species.
| Gene Used |
Target Plant |
Gene Modification Method |
Impact on Transgenic Plants |
References |
| AtKNAT7 |
Arabidopsis |
Overexpression |
Thin interfascicular fiber walls, but no change in vessel wall thickness. |
[14][13] |
| Cotton GhKNL1 |
Arabidopsis |
Overexpression |
Thinner interfascicular fibers and slightly thinner vessel walls, but no change in xylary fibers. |
[22][33] |
| Cotton GhKNAT7 |
Arabidopsis |
Overexpression |
Reduced deposition of lignocellulose in interfascicular fibers, but no change in the SCWs of xylem fibers and vessels. |
[7][24] |
| NbKNAT7 |
Tobacco |
Downregulation by VIGS and RNAi |
Increased xylem proliferation with thin-walled fiber cells, increased polysaccharide extractability, and higher saccharification rate. |
| Enhanced expression of SCW genes, CesA8, IRX9, PAL, and CCR. |
[ | 25 | ] | [17] |
| PtKNAT7 |
Poplar |
Downregulation by antisense |
Reduced expression of SCW genes, reduced lignin content, altered lignin composition (S/G ratio), and increased saccharification. |
[25][17] |
The successful complementation of
Arabidopsis knat7 Arabidopsis knat7 mutants with the overexpression of the cotton
GhKNL1 GhKNL1 gene
[22][33] and poplar
PtKNAT7 [14][13] rescued the defective
irx irx phenotype of the
knat7 knat7 mutants, suggesting the functional conservation of
KNAT7 KNAT7 genes among Arabidopsis, cotton, and poplar. The overexpression of cotton
GhKNL1 GhKNL1 in
Arabidopsis Arabidopsis resulted in thinner interfascicular fibers and slightly thinner vessels walls without any change in the xylary fibers compared to control plants
[22][33]. The overexpression of cotton
GhKNAT7 GhKNAT7 significantly reduced the deposition of lignocellulose in the interfascicular fibers of
Arabidopsis [24]. ArabidopsisHowever, the SCWs of the xylem fibers and [7].vessels However, the SCWs of the xylem fibers and vessels in the transgenic plants did not show any difference from the control plants. The dominant repression of the same cotton KNAT7 orthologue in Arabidopsis produced thinner interfascicular fibers, but thicker vessels and xylary fiber walls, suggesting that KNAT7 can act as a negative or positive regulator of SCW formation in different cell types.
Iin
the tr
esearchers' laboratoansgenic plants did not show any difference from the control plants. The dominant repression of the same cotton KNAT7 or
ythologue in Arabidopsis produced thinner interfascicular fibers,
but researchers genethicker vessels and xylary fiber walls, suggesting that KNAT7 can act as a negative or positive regulator of SCW formation in different cell types.
In our
la
ted RNAi boratory, we generated RNAi lines of tobacco (
N. benthamiana) that exhibited reduced expression of
KNAT7 [15] KNAT7 [14].
NbKNAT7 NbKNAT7 downregulated through a transient virus-induced gene silencing (VIGS) system resulted in increased xylem proliferation with thin-walled fiber cells. The glycome analyses of the cell walls showed increased polysaccharide extractability in 1 M KOH extracts of the VIGS-
NbKNAT7 NbKNAT7 lines, suggestive of SCW loosening. In addition, there were increased saccharification rates (40% higher than control) in stems of VIGS-
NbKNAT7 NbKNAT7 lines, which indicated the alteration of cell wall composition in VIGS lines downregulated for the
NbKNAT7 NbKNAT7 gene. Similar to the VIGS results, the stems of stable
RNAi RNAi lines also showed increased xylem area in their stems as compared to control stems
[15][14]. The cell walls of xylem fibers were thinner (over 50%) in the
RNAi RNAi lines as compared to vector control lines. The stems of
KNAT7 KNAT7 repression lines in tobacco showed reduced expression of SCW genes that resulted in thinner fiber cell walls with altered cell wall composition
[15][14]. All these results suggested that KNAT7 TF might act as a positive regulator of SCW formation in tobacco.
In a recent study performed in
our
esearchers' laboratory by Ahlawat et al.
[25][17], transgenic poplar plants overexpressing
PtKNAT7 PtKNAT7 and
AtKNAT7 AtKNAT7 genes showed enhanced expression of the SCW genes
CesA8, IRX9, PAL, CesA8, IRX9, PAL, and
CCR, CCR, and reduced expression of the same genes in the poplar
PtKNAT7 PtKNAT7 antisense plants. These results further suggested a positive regulatory role of KNAT7 in SCW formation in poplars. In addition, the genetic suppression of
KNAT7 KNAT7 in transgenic poplar stems reduced lignin content by about 6% and altered the lignin composition (S/G ratio) of poplar wood with increased saccharification ability (44–53% higher saccharification efficiency over control plants). Yoo et al.
[36] also reported a negative correlation between lignin content and the saccharification efficiency [26]of woody tissues and a positive correlation between the S/G ratio and the saccharification efficiency of SCW biomass. Therefore, a change in the S/G ratio and reduction in lignin content might be important for improving the saccharification efficiency of SCW biomass. All the studies reported so far in Arabidopsis and other higher plants suggest that KNAT7 acts differentially as a negative and positive regulator of SCW biosynthesis in different cell types of the same plant or in different plant also reported a negative correlation between lignin content and the saccharification efficiency of woody tissues and a positive correlation between the S/G ratio and the saccharification efficiency of SCW biomass. Therefore, a change in the S/G ratio and reduction in lignin content might be important for improving the saccharification efficiency of SCW biomass. All the studies reported so far in Arabidopsis and other higher plants suggest that KNAT7 acts differentially as a negative and positive regulator of SCW biosynthesis in different cell types of the same plant or in different plant species.species.
5. Transcriptional Network of the Class II Class II KNOXKNOX Genes Involved in SCW Formation
A complex network of transcription factors regulates SCW biosynthesis in plants
[27][28][29][30][31][32][8,9,37,38,39,40]. Among these, some Class II KNOX TFs also regulate SCW biogenesis. The major constituents of the SCW are cellulose, lignin, and hemicelluloses
[33][6]. Cellulose is a polymer of glucose synthesized at the plasma membrane by the cellulose synthase (CesA) complex
[34][41], while lignin is composed of guaiacyl (G), syringyl (S), and p-hydroxyphenyl (H) units that are synthesized through the phenylpropanoid pathway
[35][42]. Xylan is the major hemicellulose component in the SCW and consists of a linear backbone of β-(1–4)-linked D-xylosyl (Xyl) residues and α-linked (OMe(methyl)) glucuronic acid (GlcA) side branches
[36][43]. Many specific genes involved in cellulose, hemicellulose, and lignin biosynthesis pathways have previously been identified in plants (e.g.,
[36][37][38][43,44,45]) and it was anticipated that Class II KNOX TF proteins might directly regulate the expression of some of these genes. The first direct evidence of KNAT7-mediated regulation of xylan biosynthesis in the SCW was reported only recently by He et al.
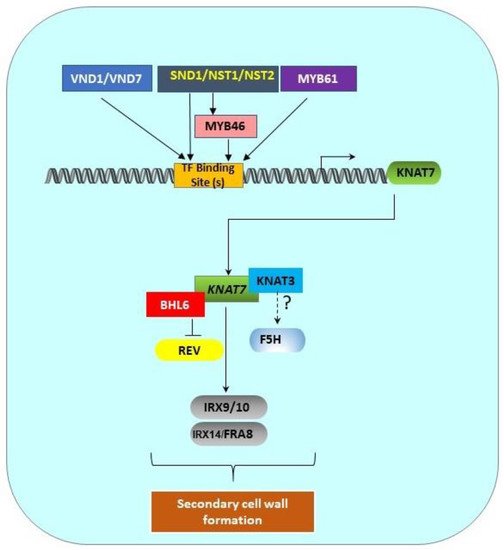
[19][31], who demonstrated that KNAT7 physically binds to the promoters of the xylan biosynthetic genes,
IRREGULAR XYLEM 9 IRREGULAR XYLEM 9 (
IRX9)
, IRX10, IRX14L, and
FRAGILE FIBER 8 (FRA8 FRAGILE FIBER 8 (FRA8;
Figure 2). Wang et al.
[39][46] also reported the involvement of KNAT7 in xylan synthesis during mucilage production. While various cellulose and lignin biosynthesis genes have been shown to be differentially expressed in various
knat7 knat7 mutants and during the ectopic expression of the
KNAT7 KNAT7 gene in transgenic plants, the direct regulation of any of these SCW genes by KNAT7 TF has not yet been reported. In addition, no information is currently available on transcriptional regulation by the TFs encoded by the other three
Class II KNOX Class II KNOX genes, namely
KNAT3 KNAT3,
KNAT4 KNAT4, and
KNAT5, KNAT5, or their orthologs in any other plant species.
Figure 2. Transcriptional regulation pathway of KNAT7 gene. SCW-associated upstream transcription factors (MYB61, SND1/NST1/NST2, VND1/VND7) and MYB46 directly bind the binding sites in the KNAT7 gene promoter to regulate the expression of the KNAT7 gene. KNAT7 positively regulates the expression of various xylan synthesis genes (IRX9/10 and IRX14L/FRA8). Interactions between KNAT7 and KNAT3 TFs might regulate F5H expression, and the interactions between KNAT7 and BLH6 negatively regulate the expression of the homeodomain-ZIP (HD-ZIP) TF gene Revoluta. All these interactions ultimately regulate SCW formation in higher plants. All genes are shown as rounded rectangles and proteins are indicated by rectangles.
Transcriptional regulation pathway of KNAT7 gene. SCW-associated upstream transcription factors (MYB61, SND1/NST1/NST2, VND1/VND7) and MYB46 directly bind the binding sites in the KNAT7 gene promoter to regulate the expression of the KNAT7 gene. KNAT7 positively regulates the expression of various xylan synthesis genes (IRX9/10 and IRX14L/FRA8). Interactions between KNAT7 and KNAT3 TFs might regulate F5H expression, and the interactions between KNAT7 and BLH6 negatively regulate the expression of the homeodomain-ZIP (HD-ZIP) TF gene Revoluta. All these interactions ultimately regulate SCW formation in higher plants. All genes are shown as rounded rectangles and proteins are indicated by rectangles.