The use of dyes dates to ancient times and has increased due to population and industrial growth, leading to the rise of synthetic dyes. These pollutants are of great environmental impact and azo dyes deserve special attention due their widespread use and challenging degradation. Among the biological solutions developed to mitigate this issue, bacteria are highlighted for being versatile organisms, which can be applied as single organism cultures, microbial consortia, in bioreactors, acting in the detoxification of azo dyes breakage by-products and have the potential to combine biodegradation with the production of products of economic interest. These characteristics go hand in hand with the ability of various strains to act under various chemical and physical parameters, such as a wide range of pH, salinity, and temperature, with good performance under industry, and environmental, relevant conditions.
- sustainability
- effluent treatment
- dyes
- bioremediation
- bacteria
- wastewater
- textile
- BES
- bioreactor
1. Introduction
2. Bacteria in the Bioremediation of Azo Dyes
Biotechnology has been widely employed in the search for solutions to the degradation and elimination of dyes, mainly because biological solutions are effective and generate less negative impact on the environment [8]. When dealing with biological processes using bacteria, especially potentially pathogenic genera and species, the concern with a possible biological impact of them, when introduced into the environment for the bioremediation process, may arise. To attend to any unwanted negative effects, some strategies can be used, some of those include: (1) the use of isolated and purified enzymes or other bacterial products that act on the discoloration without needing the bacterial cell itself [9], (2) microbial bacteria/consortia isolated from the contaminated environment itself or similar environments, in order to increase the chance of integration of the bacteria with the environment and the existing microbiota [10], (3) application of genetic engineering techniques that can develop bacterial strains with programmed death, stopping bacterial metabolism in the absence of the target contaminant [11]. Some biological bioremediation systems also have the potential of generating more than one product, in addition of decolorization, following the example of bioelectrochemical systems (BES), which helps in mitigating the costs associated with biological processes [12]. Among these solutions there is bioremediation by heterotrophic bacteria, which have, more broadly, two mechanisms related to the degradation of dyes: biosorption and enzymatic action [13].2.1. Bacterial Mechanisms of Azo Dye Degradation
2.1.1. Biosorption
2.1.2. Enzymatic Degradation
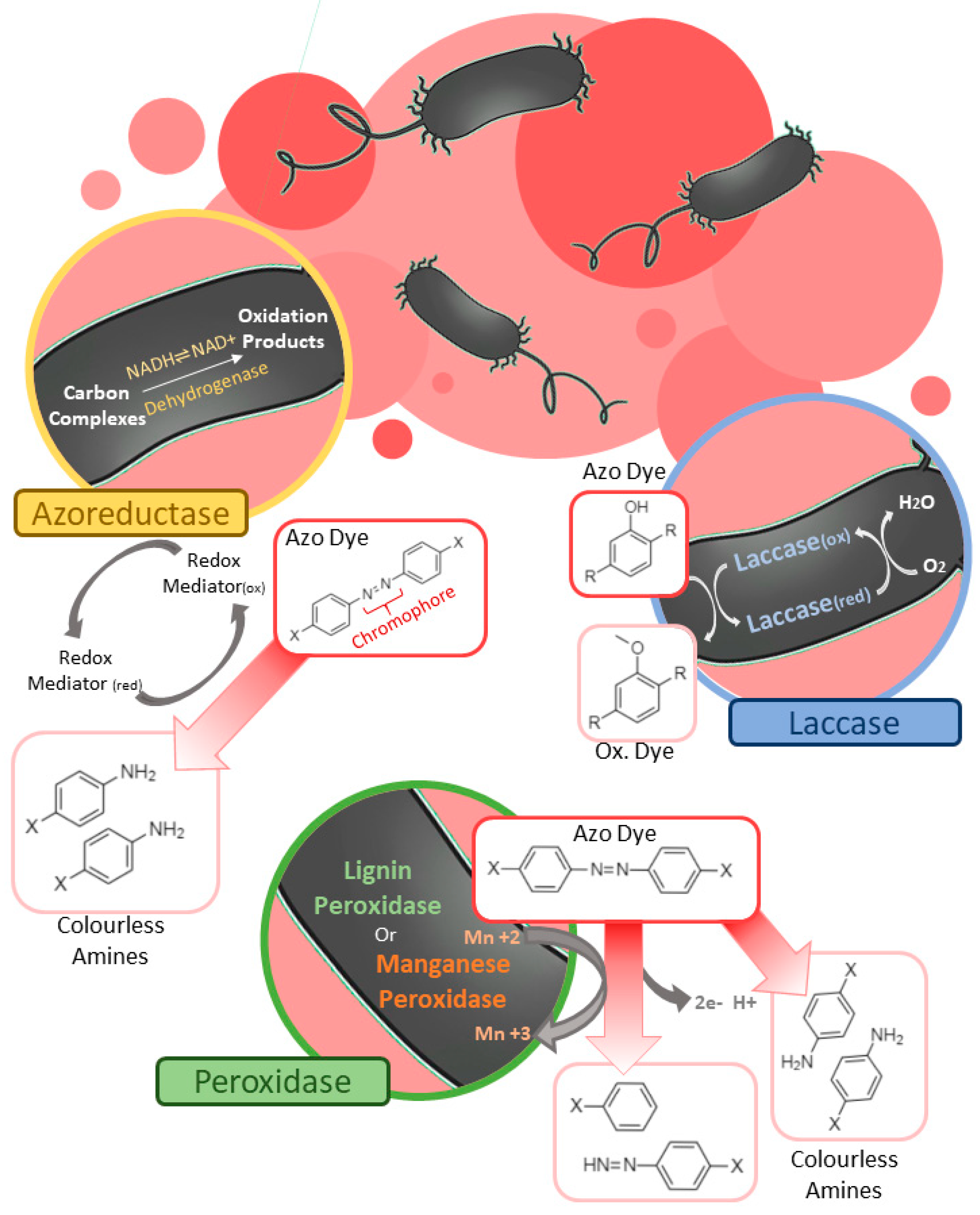
2.1.3. Enzymatic Degradation by Azoreductases
2.1.4. Enzymatic Degradation by Laccases
2.1.5. Enzymatic Degradation by Peroxidase
2.2. Bacterial Degradation of Commercial Colorants
The occurrence of bacteria in different environments and physicochemical conditions makes them an interesting focus of prospection (Table 1). In the case of dyes degradation, a wide range of variables has already been explored and it was identified that this group of microorganisms can degrade azo dyes under aerobic, microaerophilic, and anaerobic conditions, as isolated cultures or as microbial consortia, in the presence of various sources of carbon and nitrogen and in wide ranges of pH, temperature, salinity and other physical-chemical parameters. In addition, bioreactors have been used in several works in an attempt to increase the efficiency of the degradation process, especially by immobilization of microorganism or redox mediators [27].| Species | Dye | Optimum Values of Phisicochemical Parameters for Bacterial Decolorization | Degradation Mechanism | Local of Bacterial Isolation | Maximum Degradation | Reference |
|---|---|---|---|---|---|---|
| Shewanella marisflavi | Xylidine Ponceau 2R | |||||
| - | ||||||
| 89% (30 °C, 50 mg/L of dye concentration, 24h incubation and under agitation conditions) | ||||||
| [ | ||||||
| 47 | ||||||
| ] | ||||||
2.3. Degradation of Environmental and Industrial Samples
The use of bacteria to treat contamination caused by azo dyes can aim at both the treatment of effluents before their release into nature and the bioremediation of already contaminated natural environments. This topic deals with research conducted on the treatment of samples taken from contaminated environments and industrial effluents (Table 2) to show how efficient bacteria can be applied in remediating real samples in real cases of contamination.| Main Bacteria | Wastewater Source | Degradation Mechanism | Country | Maximum Degradation and Experiment Conditions | Reference | |
|---|---|---|---|---|---|---|
| Micrococcus luteus | ||||||
| 20–30% of salinity | Flocculation and Enzymatic | China | ||||
| Dyehouse | Adsorption and Enzymatic | Japan | ||||
| ≈ | 100% (30% of salinity, anaerobic conditions and 22h incubation) | [ | 28 | ] | ||
| Laboratory | [ | 48 | ] | |||
| Pseudomonas extremorientalis | Congo Red | 50 mg/L of dye concentration, 2.5–5% of salinity and 0.6 U/mL enzyme concentration | Enzymatic-Laccase | Tunísia | 79.8 ± 2.1% (50 mg/L of dye concentration, 2.5–5% of salinity, 24h incubation and 0.6 U/mL enzyme concentration) | |
| Pseudomonas aeruginosa | - | Enzymatic-Azoreductase | India | 62%-Laboratory | ||
| [ | 29 | ] | ||||
| [ | ||||||
| Aliiglaciecola lipolytica | Congo Red | 35 °C, <100 mg/L of dye concentration, 0–1% of salinity, pH 6–7, >4 g/L of glucose. | Adsorption and Enzymatic-Laccase and Azoreductase | - | >90% (35 °C, 25 mg/L of dye concentration, 1% of salinity, pH 6 and 4 g/L of glucose) | [30] |
| 49 | ] | |||||
| Pseudomonas sp. | Textile Industries | Enzymatic-Laccase | India | 90%-Laboratory | [50] | |
| Enterococcus faecalis, Shewanella indica, Oceanimonas smirnovii and Clostridium bufermentans | 8 different dyes | Varied depending of bacteria strain and dye | Enzymatic-Azoreductase and phenol oxidases | |||
| Pseudomonas sp. and | ||||||
| China | ||||||
| Bacillus sp. | ||||||
| 96.5% ( | ||||||
| Mill effluent outlet | ||||||
| E. faecalis | strain and | C | ||||
| - | India | Pseudomonas | 95% | Bacillus 97%-Laboratory | ||
| . | bufermentans | with Dye Acid Orange 7 when pH ranged from 5 to 8, respectively) | [ | 31 | ] | |
| [ | 51 | ] | ||||
| Bacillus sp. | 7 different dyes | |||||
| Pseudomonas aeruginosa, Pseudomonas putida | ||||||
| 50–100 mg/L of dye concentration, pH 10, 30 °C, with glucose and yeast extract supplementation. | ||||||
| Enzymatic | ||||||
| and | ||||||
| Ethiopia | ||||||
| Bacillus cereus | ||||||
| 100% (pH 10, 30 °C, anoxic and anaerobic conditions) | [ | |||||
| Textile Factory | ||||||
| 32 | ] | |||||
| - | Egypt | 92%-Laboratory | [ | 52 | ||
| Aeromonas hydrophila | Reactive Red 198 e Reactive Black 5 | pH 5.5–10.0, temperature were and 20–35 °C under anoxic culture | Adsorption and Enzymatic | Taiwan | >90% (pH 5.5–10.0, temperature were and 20–35 °C under anoxic culture) | [33] |
| ] | ||||||
| - | Dye Wastewater Plant | - | Korea | 75%-Real production facility | [53] | |
| Comamonas sp. | Direct Red 5B | pH 6.5, 40 °C, static incubation conditions and 300–1100 mg/L of dye concentration. | Enzymatic-Laccase and Lignin Peroxidase | India | 100% (pH 6.5, 40 °C and static incubation conditions) | [34] |
| Halomnas sp. | Remazol Black B | Varied depending of bacteria strain. | - | Iran | ≈100% (40 °C) | [35] |
| Aeromonas sp. | Reactive Black | Microaerophilic conditions | - | India | ≈100% (Microaerophilic conditions) | [36] |
| Oerskovia paurometabola | Acid Red 14 | Anaerobic conditions | Enzymatic | Portugal | 91% (anaerobic conditions) | [37] |
| Aeromonas hydrophila, Lysinibacillus sphaericus | Reactive Red 195 | - | Enzymatic-Laccase and Azoreductase | India | 91.96% (pH 8, 37 °C, 100 mg/L of dye concentration and sequential aerobic-microaerophilic conditions) | [38] |
| Bacillus sp. | 4 different dyes | - | Enzymatic-Azoreductase | - | - | [39] |
| Bacillus sp. | 5 different dyes | - | Enzymatic-Azoreductase | - | - | [40] |
| Aeromonas hydrophila, Lysinibacillus sphaericus | 5 different dyes | - | Enzymatic-Azoreductase and Laccase | India | 90.4% (pH 8, 37 °C, 100 mg/L of dye concentration and sequential aerobic-microaerophilic conditions) | [41] |
| Lysinibacillus fusiformis | Methyl Red | pH 7.5–8, 30 °C, 100 mg/L of dye concentration and 10–20% (v/v) of inoculum size | Enzymatic-Laccase, Azoreductase and Lignin Peroxidase | - | 96% (aerobic condition, pH 7.5, 30 ± 2 °C, dye concentration of 100 mg/L and 10% (v/v) inoculum size) | [42] |
| Pseudomonas stutzeri | Acid Blue 113 | - | Enzymatic-Azoreductase and Laccase | India | 86.2% (static conditions, 37 °C and 300 ppm of dye) | [43] |
| Aeromonas sp. | Methyl Orange | pH 6, 5–45 °C, 100–200 mg/L of dye concentration | Enzymatic-laccase, NADH-DCIP reductase, and azo reductase | China | ≈100% (100–200 mg/L of dye concentration; with carbon and nitrogen supplementation; pH 6; 5–45 °C) | [44] |
| Proteus mirabilis | Reactive Blue 13 | pH 7, 35 °C and anoxic conditions. | Enzymatic-Laccase, azoreductase and veratryl alcohol oxidase | Nigeria | ≈90% (pH 7) | [45] |
| Pseudomonas putida, Bacillus subtilis | 18 different dyes | - | Enzymatic-Azoreductase and Laccase | - | ≈100% | [46] |
| Bacillus sp. | Red HE7B | - | Enzymatic-Azoreductase and Laccase |
3. Conclusions
Azo dyes can be harmful to the environment and human health when disposed of without prior treatment, and the search for sustainable and less harmful production processes requires the development of new alternatives for effluent treatment that are efficient, cost-effective and of low environmental impact. Thus, bacterial bioremediation is a good alternative, given the versatility of this phylum that offers a range of possibilities, either with pure cultures or in consortia, tolerating different physicochemical parameters, in order to better adapt this process to various industrial wastes. The application of these organisms in BES also brings the possibility of generating more than one salable product or service, making this process more attractive in terms of cost, an important bottleneck to be overcome in the implementation of biological systems. The application of bacteria to environmental samples also attests to this viability, being able to degrade dyes and their toxic by-products in environmentally relevant concentrations. Through the critical reading of the literature presented, scientific advances in this area can be evaluated, as well as the efforts to remedy the still deficient points, showing bacterial bioremediation to be an increasingly feasible process. For the widespread application of bacterial bioremediation, several factors have to be considered, depending on the technique used, the characteristics of the environment to be remediated and of the bacteria strain, in this sense, the following points are relevant bottlenecks for large-scale application: (1) Bioreactor implementation and maintenance costs, (2) physicochemical parameters—which may vary over time, (3) space available for use of, e.g., wetlands or bioreactors, (4) availability of nutrients in the environment or in the textile effluent to be decontaminated, (5) presence/generation of suitable redox mediators for the enzymatic action of azo bond breaking, (6) engineering optimization in the transition from laboratory/pilot to industrial scale, (7) stricter local legislation forcing companies to treat their effluents properly, (8) co-relation between dye and bacteria/bacterial consortia or the presence of mixed dyes that can affect the bleaching given the bacterial suitability to each dye, (9) the use of industrial chemicals not considered in the laboratory tests, (10) changes in industrial dyeing techniques that modify the characteristics of its effluent and require adaptation of the bioremediation technique used, and (11) generation of toxic by-products that bacteria are not able to degradeReferences
- De Araújo, M.E.M. Corantes naturais para têxteis—Da antiguidade aos tempos modernos. Conserv. Patrim. 2006, 3–4, 39–51.
- Kant, R. Textile Dyeing Industry an Environmental Hazard. Nat. Sci. 2012, 4, 22–26.
- Abel, A. The history of dyes and pigments: From natural dyes to high performance pigments. In Colour Design: Theories and Applications; Woodhead Publishing: Sawston, UK, 2012; pp. 557–587.
- Ajaz, M.; Shakeel, S.; Rehman, A. Microbial Use for Azo Dye Degradation—A Strategy for Dye Bioremediation. Int. Microbiol. 2020, 23, 149–159.
- Guaratini, C.C.I.; Zanoni, M.V.B. Corantes Têxteis. Química Nova 2000, 23, 71–78.
- Saxena, A.; Gupta, S. Bioefficacies of Microbes for Mitigation of Azo Dyes in Textile Industry Effluent: A Review. BioResources 2020, 15, 9858–9881.
- Chequer, F.M.D.; Lizier, T.M.; de Felício, R.; Zanoni, M.V.B.; Debonsi, H.M.; Lopes, N.P.; Marcos, R.; de Oliveira, D.P. Analyses of the Genotoxic and Mutagenic Potential of the Products Formed after the Biotransformation of the Azo Dye Disperse Red 1. Toxicol. In Vitr. 2011, 25, 2054–2063.
- Singh, A.L.; Chaudhary, S.; Kayastha, A.M.; Yadav, A. Decolorization and degradation of textile effluent with the help of Enterobacter asburiae. Indian J. Biotechonol. 2015, 14, 101–106.
- Buthelezi, S.; Olaniran, A.; Pillay, B. Textile Dye Removal from Wastewater Effluents Using Bioflocculants Produced by Indigenous Bacterial Isolates. Molecules 2012, 17, 14260–14274.
- Sriram, N.; Reetha, D.; Saranraj, P. Biological Degradation of Reactive Dyes by Using Bacteria Isolated from Dye Effluent Contaminated Soil. Middle–East J. Sci. Res. 2013, 12, 1695–1700.
- Li, Q.; Wu, Y.-J. A Fluorescent, Genetically Engineered Microorganism That Degrades Organophosphates and Commits Suicide When Required. Appl. Microbiol. Biotechnol. 2009, 82, 749–756.
- Pan, Y.; Zhu, T.; He, Z. Enhanced Removal of Azo Dye by a Bioelectrochemical System Integrated with a Membrane Biofilm Reactor. Ind. Eng. Chem. Res. 2018, 57, 16433–16441.
- Kuhad, R.C.; Sood, N.; Tripathi, K.K.; Singh, A.; Ward, O.P. Developments in Microbial Methods for the Treatment of Dye Effluents. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2004; Volume 56, pp. 185–213. ISBN 9780120026586.
- Solís, M.; Solís, A.; Pérez, H.I.; Manjarrez, N.; Flores, M. Microbial Decolouration of Azo Dyes: A Review. Process Biochem. 2012, 47, 1723–1748.
- Khan, R.; Bhawana, P.; Fulekar, M.H. Microbial Decolorization and Degradation of Synthetic Dyes: A Review. Rev. Environ. Sci. Biotechnol. 2013, 12, 75–97.
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S.P. Bacterial Decolorization and Degradation of Azo Dyes: A Review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157.
- Singh, R.L.; Singh, P.K.; Singh, R.P. Enzymatic Decolorization and Degradation of Azo Dyes—A Review. Int. Biodeterior. Biodegrad. 2015, 104, 21–31.
- Keck, A.; Klein, J.; Kudlich, M.; Stolz, A.; Knackmuss, H.J.; Mattes, R. Reduction of Azo Dyes by Redox Mediators Originating in the Naphthalenesulfonic Acid Degradation Pathway of Sphingomonas Sp. Strain BN6. Appl. Environ. Microbiol. 1997, 63, 3684–3690.
- Chivukula, M.; Renganathan, V. Phenolic Azo Dye Oxidation by Laccase from Pyricularia Oryzae. Appl. Environ. Microbiol. 1995, 61, 4374–4377.
- Garcia, F.D.S. Enzimas Oxidorredutases Produzidas por Fungos Filamentosos. Master’s Thesis, Universidade de São Paulo, São Paulo, Brasil, 13 June 2018.
- Chacko, J.T.; Subramaniam, K. Enzymatic degradation of azo dyes-a review. Int. J. Environ. Sci. 2011, 1, 1250, 2011.
- Misal, S.A.; Gawai, K.R. Azoreductase: A Key Player of Xenobiotic Metabolism. Bioresour. Bioprocess. 2018, 5, 17.
- Sharma, V.; Upadhyay, L.S.B.; Vasanth, D. Extracellular Thermostable Laccase-Like Enzymes from Bacillus licheniformis Strains: Production, Purification and Characterization. Appl. Biochem. Microbiol. 2020, 56, 420–432.
- Kandelbauer, A.; Guebitz, G.M. Bioremediation for the Decolorization of Textile Dyes—A Review. In Environmental Chemistry; Lichtfouse, E., Schwarzbauer, J., Robert, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 269–288. ISBN 9783540228608.
- Paszczynski, A.; Crawford, R.L. Degradation of Azo Compounds by Ligninase from Phanerochaete Chrysosporium: Involvement of Veratryl Alcohol. Biochem. Biophys. Res. Commun. 1991, 178, 1056–1063.
- Ilić Đurđić, K.; Ostafe, R.; Đurđević Đelmaš, A.; Popović, N.; Schillberg, S.; Fischer, R.; Prodanović, R. Saturation Mutagenesis to Improve the Degradation of Azo Dyes by Versatile Peroxidase and Application in Form of VP-Coated Yeast Cell Walls. Enzym. Microb. Technol. 2020, 136, 109509.
- Singh, P.; Iyengar, L.; Pandey, A. Bacterial Decolorization and Degradation of Azo Dyes. In Microbial Degradation of Xenobiotics; Singh, S.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 101–133. ISBN 9783642237881.
- Xu, F.; Mou, Z.; Geng, J.; Zhang, X.; Li, C. Azo Dye Decolorization by a Halotolerant Exoelectrogenic Decolorizer Isolated from Marine Sediment. Chemosphere 2016, 158, 30–36.
- Neifar, M.; Chouchane, H.; Mahjoubi, M.; Jaouani, A.; Cherif, A. Pseudomonas extremorientalis BU118: A New Salt-Tolerant Laccase-Secreting Bacterium with Biotechnological Potential in Textile Azo Dye Decolourization. 3 Biotech 2016, 6, 107.
- Wang, Y.; Jiang, L.; Shang, H.; Li, Q.; Zhou, W. Treatment of Azo Dye Wastewater by the Self-Flocculating Marine Bacterium Aliiglaciecola Lipolytica. Environ. Technol. Innov. 2020, 19, 100810.
- Zhuang, M.; Sanganyado, E.; Zhang, X.; Xu, L.; Zhu, J.; Liu, W.; Song, H. Azo Dye Degrading Bacteria Tolerant to Extreme Conditions Inhabit Nearshore Ecosystems: Optimization and Degradation Pathways. J. Environ. Manag. 2020, 261, 110222.
- Guadie, A.; Tizazu, S.; Melese, M.; Guo, W.; Ngo, H.H.; Xia, S. Biodecolorization of Textile Azo Dye Using Bacillus sp. Strain CH12 Isolated from Alkaline Lake. Biotechnol. Rep. 2017, 15, 92–100.
- Chen, K.-C.; Wu, J.-Y.; Liou, D.-J.; Hwang, S.-C.J. Decolorization of the Textile Dyes by Newly Isolated Bacterial Strains. J. Biotechnol. 2003, 101, 57–68.
- Jadhav, U.U.; Dawkar, V.V.; Ghodake, G.S.; Govindwar, S.P. Biodegradation of Direct Red 5B, a Textile Dye by Newly Isolated Comamonas Sp. UVS. J. Hazard. Mater. 2008, 158, 507–516.
- Asad, S.; Amoozegar, M.A.; Pourbabaee, A.A.; Sarbolouki, M.N.; Dastgheib, S.M.M. Decolorization of Textile Azo Dyes by Newly Isolated Halophilic and Halotolerant Bacteria. Bioresour. Technol. 2007, 98, 2082–2088.
- Shah, M. Evaluation of Aeromonas Spp. In Microbial Degradation and Decolorization of Reactive Black in Microaerophilic—Aerobic Condition. J. Bioremed. Biodeg. 2014, 5, 246.
- Franca, R.D.G.; Vieira, A.; Carvalho, G.; Oehmen, A.; Pinheiro, H.M.; Barreto Crespo, M.T.; Lourenço, N.D. Oerskovia Paurometabola Can Efficiently Decolorize Azo Dye Acid Red 14 and Remove Its Recalcitrant Metabolite. Ecotoxicol. Environ. Saf. 2020, 191, 110007.
- Srinivasan, S.; Sadasivam, S.K. Exploring Docking and Aerobic-Microaerophilic Biodegradation of Textile Azo Dye by Bacterial Systems. J. Water Process Eng. 2018, 22, 180–191.
- Dehghanian, F.; Kay, M.; Kahrizi, D. A Novel Recombinant AzrC Protein Proposed by Molecular Docking and in Silico Analyses to Improve Azo Dye’s Binding Affinity. Gene 2015, 569, 233–238.
- Haghshenas, H.; Kay, M.; Dehghanian, F.; Tavakol, H. Molecular Dynamics Study of Biodegradation of Azo Dyes via Their Interactions with AzrC Azoreductase. J. Biomol. Struct. Dyn. 2016, 34, 453–462.
- Srinivasan, S.; Shanmugam, G.; Surwase, S.V.; Jadhav, J.P.; Sadasivam, S.K. In Silico Analysis of Bacterial Systems for Textile Azo Dye Decolorization and Affirmation with Wetlab Studies: General. Clean Soil Air Water 2017, 45, 1600734.
- Sari, I.P.; Simarani, K. Decolorization of Selected Azo Dye by Lysinibacillus fusiformis W1B6: Biodegradation Optimization, Isotherm, and Kinetic Study Biosorption Mechanism. Adsorpt. Sci. Technol. 2019, 37, 492–508.
- Joshi, A.U.; Hinsu, A.T.; Kotadiya, R.J.; Rank, J.K.; Andharia, K.N.; Kothari, R.K. Decolorization and Biodegradation of Textile Di-Azo Dye Acid Blue 113 by Pseudomonas stutzeri AK6. 3 Biotech 2020, 10, 214.
- Du, L.-N.; Li, G.; Zhao, Y.-H.; Xu, H.-K.; Wang, Y.; Zhou, Y.; Wang, L. Efficient Metabolism of the Azo Dye Methyl Orange by Aeromonas Sp. Strain DH-6: Characteristics and Partial Mechanism. Int. Biodeterior. Biodegrad. 2015, 105, 66–72.
- Olukanni, O.D.; Osuntoki, A.A.; Kalyani, D.C.; Gbenle, G.O.; Govindwar, S.P. Decolorization and Biodegradation of Reactive Blue 13 by Proteus Mirabilis LAG. J. Hazard. Mater. 2010, 184, 290–298.
- Mendes, S.; Farinha, A.; Ramos, C.G.; Leitão, J.H.; Viegas, C.A.; Martins, L.O. Synergistic Action of Azoreductase and Laccase Leads to Maximal Decolourization and Detoxification of Model Dye-Containing Wastewaters. Bioresour. Technol. 2011, 102, 9852–9859.
- Thakur, J.K.; Paul, S.; Dureja, P.; Annapurna, K.; Padaria, J.C.; Gopal, M. Degradation of Sulphonated Azo Dye Red HE7B by Bacillus sp. and Elucidation of Degradative Pathways. Curr. Microbiol. 2014, 69, 183–191.
- Ito, T.; Shimada, Y.; Suto, T. Potential Use of Bacteria Collected from Human Hands for Textile Dye Decolorization. Water Resour. Ind. 2018, 20, 46–53.
- Sarayu, K.; Sandhya, S. Aerobic Biodegradation Pathway for Remazol Orange by Pseudomonas aeruginosa. Appl. Biochem. Biotechnol. 2010, 160, 1241–1253.
- Telke, A.A.; Joshi, S.M.; Jadhav, S.U.; Tamboli, D.P.; Govindwar, S.P. Decolorization and Detoxification of Congo Red and Textile Industry Effluent by an Isolated Bacterium Pseudomonas sp. SU-EBT. Biodegradation 2010, 21, 283–296.
- Srinivasan, V.; Bhavan, P.S.; Krishnakumar, J. Bioremediation of textile dye effluent by Bacillus and Pseudomonas spp. Int. J. Environ. Sci. Technol. 2014, 3, 2215–2224.
- Bayoumi, M.N.; Al-Wasify, R.S.; Hamed, S.R. Bioremediation of textile wastewater dyes using local bacterial isolates. Int. J. Curr. Microbiol. App. Sci. 2014, 3, 962–970.
- Kalathil, S.; Lee, J.; Cho, M.H. Efficient Decolorization of Real Dye Wastewater and Bioelectricity Generation Using a Novel Single Chamber Biocathode-Microbial Fuel Cell. Bioresour. Technol. 2012, 119, 22–27.
