Nervous necrosis virus (NNV) can infect many species of fish and caused an 80~100% mortality rate. NNV capsid protein (NNVCP) was the only structural proteins of NNV, but there are few studies on the protein-protein interaction between NNVCP and host cell. Here, we demonstrated 49 proteins in optic nerve tissue that could interact with NNVCP. A new mechanism of NNV morphogenesis is clarified by exploring the interaction between NNVCP and host cells. Moreover, CKB was identified as a novel enhancer for NNV through interacting with NNVCP. These findings may provide more accurate target proteins to understand NNV morphogenesis and provide new insights into the antiviral infection.
- Nervous necrosis virus
- immunoprecipitation assay
- protein-protein interaction
- creatine kinase
- far-western blot
1. Introduction
Grouper is an aquaculture fish with substantial economic value and it is an important source of income in many Asian countries. Like other high-density aquaculture species, groupers are also threatened by many pathogens, including Vibrio alginolyticus, iridovirus, and nervous necrosis virus (NNV) [1][2][3][4][1,2,3,4]. NNV is highly lethal to all grouper stages and results in massive economic losses worldwide [5]. NNV is the causative agent of viral nervous necrosis (VNN) disease, which has a wide host range, infecting at least 40 families of fish species [5][6][7][8][9][6,7,8,9,10]. NNV infects the central nervous system and causes vacuolation of the brain and retina [10][11]. NNV has a non-enveloped icosahedral structure and belongs to the family Nodaviridae (genus Betanodavirus), and its genome consists of two single-stranded positive-sense RNAs. RNA1 encodes the RNA-dependent RNA polymerase (RdRp), and RNA2 encodes the viral capsid protein [11][12][13][14][15][12,13,14,15,16]. The NNV capsid protein (NNVCP) is the only structural protein of the virion and has been shown to determine the host range [16][17]. Betanodaviruses have been classified into the following four genotypes based on the sequence of the RNA2 segment: tiger puffer NNV (TPNNV), striped jacked NNV (SJNNV), red-spotted grouper NNV (RGNNV), and barfin flounder NNV (BFNNV) [17][18]. Despite its relatively simple virion structure, no clear NNV receptor has been identified, and the mechanism of NNV infection remains unclear.
It is very difficult to prevent NNV in the aquaculture process except through preventing infection by parental fish, ensuring water quality, and actively managing feed and livestock. NNV is very contagious, and it is difficult to completely eliminate NNV from aquaculture environments. DNA vaccines, interferons, or immunostimulants can be used to reduce NNV infections and outbreaks, but they are not sufficient to completely control the epidemic [18][19][20][21][19,20,21,22]. However, the functional roles of host factors interacting with the NNVCP in viral genome replication remain ambiguous.
Although NNV can infect more than 30 marine and freshwater fish species, and furthermore, NNV capsid protein (NNVCP) was the only external viral structural protein in virus particles, to date, only GHSC70 and MmHSP90ab1 have been identified as attachment receptors for NNV [22][23][23,24]. However, preventing these proteins from interacting with cells cannot inhibit viral infection, so NNV is thought to still have other undiscovered host receptors or co-receptors. In addition, many cellular proteins that may be involved in the NNV morphogenesis have not been verified. Thus, native NNVCP isolated from the grouper cell line GF-1, rather than a recombinant protein generated by bacteria, was applied in this study to investigate the proteins that can interact with NNVCP. Optic nerves were selected from uninfected orange-spotted grouper (Epinephelus coioides) to evaluate protein interactions with native NNVCP by immunoprecipitation (IP). We annotated the proteins using 1DLC, LTQ-Orbitrap MS database of the Epinephelus genus in NCBI and UniProt. The 49 identified proteins could contribute as putative receptor or co-receptor, cytoskeleton, glucose metabolism, and ATP generation, immunity, mitochondrial ion regulation, and ribosomal proteins.
2. Virus Infection
NNV was isolated from grouper larvae with NNV disease. NNV was propagated in GF-1 cells at a multiplicity of infection (MOI) of 10, and infected cells were incubated in L-15 medium with 2% FBS at 27 °C for three days until cytopathic effect (CPE) was observed. After CPE was observed, the culture supernatant was collected and centrifuged at 12,000 rpm for 5 min at 4 °C and then kept as the virus stock. To further collect cell lysate, 2 mL of PBS were used to wash cells, and the virus-infected cells were then treated with lysis buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, 0.1% NP-40, and 0.02% protease inhibitor, pH 8). After slow shaking for 30 min at 4 °C, the cell lysate was collected in a 1.5 mL microcentrifuge tube, sonicated for 2 min, and then centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatant was collected in a new 1.5 mL microcentrifuge tube and stored at −20 °C.
3. Identification of Optic Nerve Proteins Interacting with Native NNV Capsid Protein (NNVCP) by Proteomic Analysis of Immunoprecipitation (IP) Assay
To identify host proteins interacting with native NNVCP, the protein–protein interaction assay was performed using an IP assay with natural NNVCP and grouper optic nerve tissue. Protein A beads conjugated with anti-NNVCP specific antibody were first incubated with native NNVCP produced from NNV-infected GF-1 cells and then co-incubated with lysates from grouper optic nerve tissue (Figure 1, lane 1). In the control group, Protein A beads conjugated with anti-NNVCP specific antibodies were incubated directly with the lysate of the optic nerve tissue without adding native NNVCP (Figure 1, lane 2). Three obvious bands were detected in Figure 1: 37 kDa is native NNVCP (Figure 1, lane 1 and lane 4), 25 kDa is the light chain of anti-NNVCP specific antibody, and 50 kDa is the heavy chain of anti-NNVCP specific antibody (Figure 1, lane1 and lane 2). Lane 3 revealed that grouper optic nerve was non-infected NNV. After IP, samples were preliminary evaluated by SDS-PAGE and Western blot. Through mass spectrometry (MS) analysis of optic nerve IP cleavage samples, it was identified that many grouper proteins could interact with NNVCP, and the proteins were analyzed and annotated (Table 1 and Table 2). The score in TableTable 2 2 indicates the protein score (MudPI score), which is the sum of the peptide score [24][26]. MS analysis of the grouper optic nerve protein that can interact with NNVCP found 287 peptides belonging to 49 proteins. Among the proteins involved in multiple functional categories, 12 are ribosomal proteins, 7 are involved in immunity including heat shock protein 60 and heat shock protein 90, 7 are involved in glucose metabolism and ATP generation (such as creatine kinase), 7 are related to the cytoskeleton, 5 are Ca2+ binding proteins, 2 are ion regulation proteins, 3 are involved in lipid metabolism, 2 are involved in apoptosis, and 2 are involved in protein hydrolysis. The functions of hemoglobin α chain and Hnrpa01 protein are not very clear (Table 1).
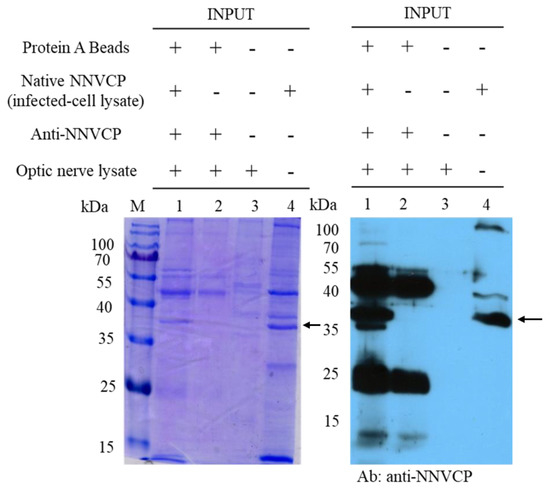
Figure 1.
Table 2. The original protein assay data in the grouper optic nerve with native NNVCP by immunoprecipitation. The gray font indicates the proteins that were also detected in the control group.
| GenBank Accession No. | Description | Score * |
|---|
| AAR97600.2 | beta actin (Epinephelus coioides) | 2492.00 | ||
| 3JBM | C Chain C, Virus-like Particle of Orange-spotted Grouper Nervous Necrosis Virus | 1706.91 | ||
| AER42656.1 | keratin 8, partial (Epinephelus coioides) | 1457.49 | ||
| AAX78203.1 | immunoglobulin mu heavy chain ( | Epinephelus coioides | ) | 1263.15 |
| AAX78206.1 | immunoglobulin mu heavy chain ( | Epinephelus coioides | ) | 999.30 |
| AGG55392.1 | voltage-dependent anion selective channel protein 2 ( | Epinephelus coioides | ) | 944.17 |
| AAX78208.1 | immunoglobulin mu heavy chain ( | Epinephelus coioides | ) | 914.30 |
| AEG78351.1 | keratin type II E3, partial ( | Epinephelus coioides | ) | 790.40 |
| ACM48181.1 | apolipoprotein AI, partial ( | Epinephelus coioides | ) | 745.80 |
| AGG55391.1 | heat shock cognate protein 70 (Epinephelus coioides) | 745.79 | ||
| ACH73075.1 | keratin 8, partial (Epinephelus coioides) | 740.04 | ||
| AIS72878.1 | heat shock protein 60 ( | Epinephelus coioides | ) | 679.74 |
| AEW43726.1 | transferrin ( | Epinephelus coioides | ) | 645.54 |
| AER42652.1 | hemoglobin beta chain (Epinephelus coioides) | 623.08 | ||
| ADG29138.1 | tropomyosin ( | Epinephelus coioides | ) | 539.59 |
| ABW74631.1 | immunoglobulin light chain ( | Epinephelus coioides | ) | 504.86 |
| AHB51756.1 | calmodulin ( | Epinephelus coioides | ) | 491.05 |
| ABW04131.1 | glyceraldehyde-3-phosphate dehydrogenase, partial ( | Epinephelus coioides | ) | 484.23 |
| AAS55942.1 | immunoglobulin light chain variable region ( | Epinephelus coioides | ) | 476.38 |
| ABW04145.1 | smooth muscle cell-specific protein SM22 alpha ( | Epinephelus coioides | ) | 430.16 |
| AEO89322.1 | voltage dependent anion channel protein 1 ( | Epinephelus coioides | ) | 421.60 |
| AER42688.1 | tropomyosin alpha-4 chain, partial ( | Epinephelus coioides | ) | 418.42 |
| ABW04135.1 | natural killer cell enhancement factor ( | Epinephelus coioides | ) | 361.05 |
| AEG78406.1 | hemoglobin alpha chain, partial ( | Epinephelus coioides | ) | 347.41 |
| ADG29126.1 | fructose-bisphosphate aldolase A ( | Epinephelus coioides | ) | 340.73 |
| AEG78409.1 | 60S ribosomal protein L30 ( | Epinephelus coioides | ) | 325.27 |
| ADG29180.1 | triosephosphate isomerase B ( | Epinephelus coioides | ) | 310.40 |
| ACL98136.1 | type I keratin, partial (Epinephelus coioides) | 305.82 | ||
| ABW04143.1 | S100-like calcium binding protein ( | Epinephelus coioides | ) | 298.82 |
| ABW04124.1 | ADP-ATP translocase ( | Epinephelus coioides | ) | 288.91 |
| ACH73065.1 | ribosomal protein S7 ( | Epinephelus coioides | ) | 283.43 |
| ADZ76534.1 | myosin light chain 3 ( | Epinephelus coioides | ) | 279.24 |
| ABW04132.1 | Hnrpa01 protein, partial ( | Epinephelus coioides | ) | 269.70 |
| ACV04938.1 | heat shock protein 90 ( | Epinephelus coioides | ) | 269.27 |
| ADG29156.1 | histone H2B (Epinephelus coioides) | 265.26 | ||
| AOW69105.1 | elongation factor 1-alpha ( | Epinephelus coioides | ) | 264.68 |
| ADG29136.1 | beta-enolase (Epinephelus coioides) | 247.11 | ||
| AEG78428.1 | 60S ribosomal protein L10a (Epinephelus coioides) | 246.47 | ||
| AAW29021.1 | lactate dehydrogenase-A, partial ( | Epinephelus coioides | ) | 212.87 |
| ABW04139.1 | ribosomal protein LP0 ( | Epinephelus coioides | ) | 212.25 |
| ABW04136.1 | nucleoside-diphosphate kinase ( | Epinephelus coioides | ) | 199.15 |
| ADG29150.1 | 60S ribosomal protein L19, partial (Epinephelus coioides) | 186.15 | ||
| ADG29176.1 | muscle phosphoglycerate mutase 2 (Epinephelus coioides) | 184.01 | ||
| ACH73061.1 | ribosomal protein L7, partial ( | Epinephelus coioides | ) | 157.36 |
| ABW04127.1 | brain-type fatty acid binding protein ( | Epinephelus coioides | ) | 156.32 |
| ABW04123.1 | 60S ribosomal protein L27 ( | Epinephelus coioides | ) | 152.44 |
| ADZ99127.1 | immunoglobulin heavy chain variable region, partial ( | Epinephelus coioides | ) | 146.61 |
| AEG78395.1 | aldehyde dehydrogenase family 9 member A1-A, partial ( | Epinephelus coioides | ) | 136.08 |
| ADG29178.1 | tryptase-2 precursor, partial ( | Epinephelus coioides | ) | 135.49 |
| ACL98142.1 | desmin, partial (Epinephelus coioides) | 120.60 | ||
| ABW04138.1 | ribosomal protein L23, partial ( | Epinephelus coioides | ) | 110.93 |
| ACH73060.1 | ictacalcin ( | Epinephelus coioides | ) | 104.23 |
| ABW04130.1 | galactoside binding lectin ( | Epinephelus coioides | ) | 100.05 |
| AER42692.1 | 60S ribosomal protein LP1 ( | Epinephelus coioides | ) | 99.41 |
| AEG78402.1 | 40S ribosomal protein S18 ( | Epinephelus coioides | ) | 87.32 |
| ADG29169.1 | 60S ribosomal protein L13, partial ( | Epinephelus coioides | ) | 86.49 |
| ADG29143.1 | 60S ribosomal protein L31 ( | Epinephelus coioides | ) | 67.76 |
| ACM41841.1 | creatine kinase, partial ( | Epinephelus coioides | ) | 58.09 |
| AEG78365.1 | 40S ribosomal protein S2 ( | Epinephelus coioides | ) | 57.41 |
| AEG78426.1 | eukaryotic translation elongation factor 2, partial ( | Epinephelus coioides | ) | 55.83 |
| ABW74647.1 | immunoglobulin light chain ( | Epinephelus coioides | ) | 51.54 |
| ACM41842.1 | 14 kDa apolipoprotein, partial ( | Epinephelus coioides | ) | 50.87 |
| ACL98134.1 | ran protein, partial ( | Epinephelus coioides | ) | 48.98 |
| AHA43788.1 | calreticulin ( | Epinephelus coioides | ) | 38.35 |
| ABW04141.1 | ribosomal protein S13, partial ( | Epinephelus coioides | ) | 32.62 |
References
- Hegde, A.; Chen, C.; Qin, Q.; Lam, T.; Sin, Y. Characterization, pathogenicity and neutralization studies of a nervous necrosis virus isolated from grouper, Epinephelus tauvina, in Singapore. Aquaculture 2002, 213, 55–72. [Google Scholar] [CrossRef]
- Qin, Q.; Chang, S.; Ngoh-Lim, G.; Gibson-Kueh, S.; Shi, C.; Lam, T. Characterization of a novel ranavirus isolated from grouper Epinephelus tauvina. Dis. Aquat. Org. 2003, 53, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Huang, Y.; Zhu, W.; Li, C.; Huang, X.; Qin, Q. Isolation and identification of Singapore grouper iridovirus Hainan strain (SGIV-HN) in China. Arch. Virol. 2019, 164, 1869–1872. [Google Scholar] [CrossRef] [PubMed]
- Bandín, I.; Souto, S. Betanodavirus and VER disease: A 30-year research review. Pathogens 2020, 9, 106. [Google Scholar] [CrossRef]
- Arimoto, M.; Maruyama, K.; Furusawa, I. Epizootiology of viral nervous necrosis (VNN) in striped jack. Fish Pathol. 1994, 29, 19–24. [Google Scholar] [CrossRef]
- Munday, B.; Kwang, J.; Moody, N. Betanodavirus infections of teleost fish: A review. J. Fish Dis. 2002, 25, 127–142. [Google Scholar] [CrossRef]
- Kara, H.; Chaoui, L.; Derbal, F.; Zaidi, R.; De Boisséson, C.; Baud, M.; Bigarré, L. Betanodavirus-associated mortalities of adult wild groupers E pinephelus marginatus (Lowe) and E pinephelus costae (Steindachner) in A lgeria. J. Fish Dis. 2014, 37, 273–278. [Google Scholar] [CrossRef]
- Moody, N.; Horwood, P.; Reynolds, A.; Mahony, T.; Anderson, I.; Oakey, H. Phylogenetic analysis of betanodavirus isolates from Australian finfish. Dis. Aquat. Org. 2009, 87, 151–160. [Google Scholar] [CrossRef]
- Vendramin, N.; Patarnello, P.; Toffan, A.; Panzarin, V.; Cappellozza, E.; Tedesco, P.; Terlizzi, A.; Terregino, C.; Cattoli, G. Viral Encephalopathy and Retinopathy in groupers (Epinephelus spp.) in southern Italy: A threat for wild endangered species? BMC Vet. Res. 2013, 9, 1–7. [Google Scholar] [CrossRef]
- Costa, J.Z.; Thompson, K.D. Understanding the interaction between Betanodavirus and its host for the development of prophylactic measures for viral encephalopathy and retinopathy. Fish Shellfish Immunol. 2016, 53, 35–49. [Google Scholar] [CrossRef]
- Chi, S.; Lo, C.; Kou, G.; Chang, P.; Peng, S.; Chen, S. Mass mortalities associated with viral nervous necrosis (VNN) disease in two species of hatchery-reared grouper, Epinephelus fuscogutatus and Epinephelus akaara (Temminck & Schlegel). J. Fish Dis. 1997, 20, 185–193. [Google Scholar]
- Mori, K.-I.; Nakai, T.; Muroga, K.; Arimoto, M.; Mushiake, K.; Furusawa, I. Properties of a new virus belonging to Nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology 1992, 187, 368–371. [Google Scholar] [CrossRef]
- Chen, L.-J.; Su, Y.-C.; Hong, J.-R. Betanodavirus non-structural protein B1: A novel anti-necrotic death factor that modulates cell death in early replication cycle in fish cells. Virology 2009, 385, 444–454. [Google Scholar] [CrossRef]
- Fenner, B.J.; Thiagarajan, R.; Chua, H.K.; Kwang, J. Betanodavirus B2 is an RNA interference antagonist that facilitates intracellular viral RNA accumulation. J. Virol. 2006, 80, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, T.; Mise, K.; Takeda, A.; Okinaka, Y.; Mori, K.-I.; Arimoto, M.; Okuno, T.; Nakai, T. Characterization of Striped jack nervous necrosis virus subgenomic RNA3 and biological activities of its encoded protein B2. J. Gen. Virol. 2005, 86, 2807–2816. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Teng, Y.; Zheng, X.; Wu, Y.; Xie, X.; He, J.; Ye, Y.; Wu, Z. Complete sequence of a viral nervous necrosis virus (NNV) isolated from red-spotted grouper (Epinephelus akaara) in China. Arch. Virol. 2012, 157, 777–782. [Google Scholar] [CrossRef]
- Iwamoto, T.; Okinaka, Y.; Mise, K.; Mori, K.-I.; Arimoto, M.; Okuno, T.; Nakai, T. Identification of host-specificity determinants in betanodaviruses by using reassortants between striped jack nervous necrosis virus and sevenband grouper nervous necrosis virus. J. Virol. 2004, 78, 1256–1262. [Google Scholar] [CrossRef]
- Nishizawa, T.; Furuhashi, M.; Nagai, T.; Nakai, T.; Muroga, K. Genomic classification of fish nodaviruses by molecular phylogenetic analysis of the coat protein gene. Appl. Environ. Microbiol. 1997, 63, 1633–1636. [Google Scholar] [CrossRef]
- Kuan, Y.C.; Sheu, F.; Lee, G.C.; Tsai, M.W.; Hung, C.L.; Nan, F.H. Administration of recombinant Reishi immunomodulatory protein (rLZ-8) diet enhances innate immune responses and elicits protection against nervous necrosis virus in grouper Epinephelus coioides. Fish Shellfish Immunol. 2012, 32, 986–993. [Google Scholar] [CrossRef]
- Wang, Y.D.; Rajanbabu, V.; Chen, J.Y. Transcriptome analysis of medaka following epinecidin-1 and TH1-5 treatment of NNV infection. Fish Shellfish Immunol. 2015, 42, 121–131. [Google Scholar] [CrossRef]
- Kuo, H.P.; Chung, C.L.; Hung, Y.F.; Lai, Y.S.; Chiou, P.P.; Lu, M.W.; Kong, Z.L. Comparison of the responses of different recombinant fish type I interferons against betanodavirus infection in grouper. Fish Shellfish Immunol. 2016, 49, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Lomonossoff, G.P.; Marsian, J.; Hurdiss, D.L.; Ranson, N.A.; Ritala, A.; Paley, R.K.; Cano Cejas, I. Plant-made nervous necrosis virus-like particles protect fish against disease. Front. Plant Sci. 2019, 10, 880. [Google Scholar]
- Chang, J.S.; Chi, S.C. GHSC70 is involved in the cellular entry of nervous necrosis virus. J. Virol. 2015, 89, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jia, K.; Jia, P.; Xiang, Y.; Lu, X.; Liu, W.; Yi, M. Marine medaka heat shock protein 90ab1 is a receptor for red-spotted grouper nervous necrosis virus and promotes virus internalization through clathrin-mediated endocytosis. PLoS Pathog. 2020, 16, e1008668. [Google Scholar] [CrossRef]
