Streptococcus suis is a pathogen of pigs that can cause infections in humans who are in close contact with infected animals and/or contaminated pork-derived products, as well as those who have consumed raw pork products. Several molecular methods have been applied to investigate S. suis strain diversity and identify phylogenetic groups. Multilocus sequence typing (MLST), commonly used to differentiate between S. suis strains, has been instrumental in identifying that the species is genetically highly diverse. Recent advances in whole-genome analysis have resulted in schemes permitting the classification of S. suis populations as pathogenic or non-pathogenic, or disease-associated or non-disease associated.
- Streptococcus suis
- clonal complex
- multilocus sequence typing (MLST)
- PCR
- minimum core genome sequence typing (MCG)
- pathotyping
1. Introduction
|
Characteristic |
WGS |
MLST |
Multiplex PCR-CC |
RAPD |
PCR-RFLP |
MLVA |
AFLP |
PFGE |
Ribotyping |
|---|---|---|---|---|---|---|---|---|---|
|
Reproducibility |
Good |
Good |
Good |
Poor to moderate |
Moderate |
Good |
Good |
Good |
Good |
|
Discriminatory power |
Excellent |
High |
Moderate |
Moderate to good |
Poor to moderate |
Excellent |
Excellent |
Excellent |
Good |
|
Ease of use |
Moderately labor-intensive |
Simple to moderate labor |
Simple |
Simple |
Simple |
Simple |
Moderate |
Labor-intensive |
Labor-intensive |
|
Interpretation |
Moderate to very complex |
Simple to moderate |
Simple |
Moderate to complex |
Simple |
Simple |
Complex |
Moderate to complex |
Moderate to complex |
|
Cost |
Very high |
Moderate |
Low |
Low |
Low |
Low to moderate |
Moderate |
High |
High |
|
Universal applicability |
Yes |
Yes |
Limit to some CCs |
Yes |
Yes |
Yes |
Yes |
Yes |
Yes |
2. Multilocus Sequence Typing (MLST)
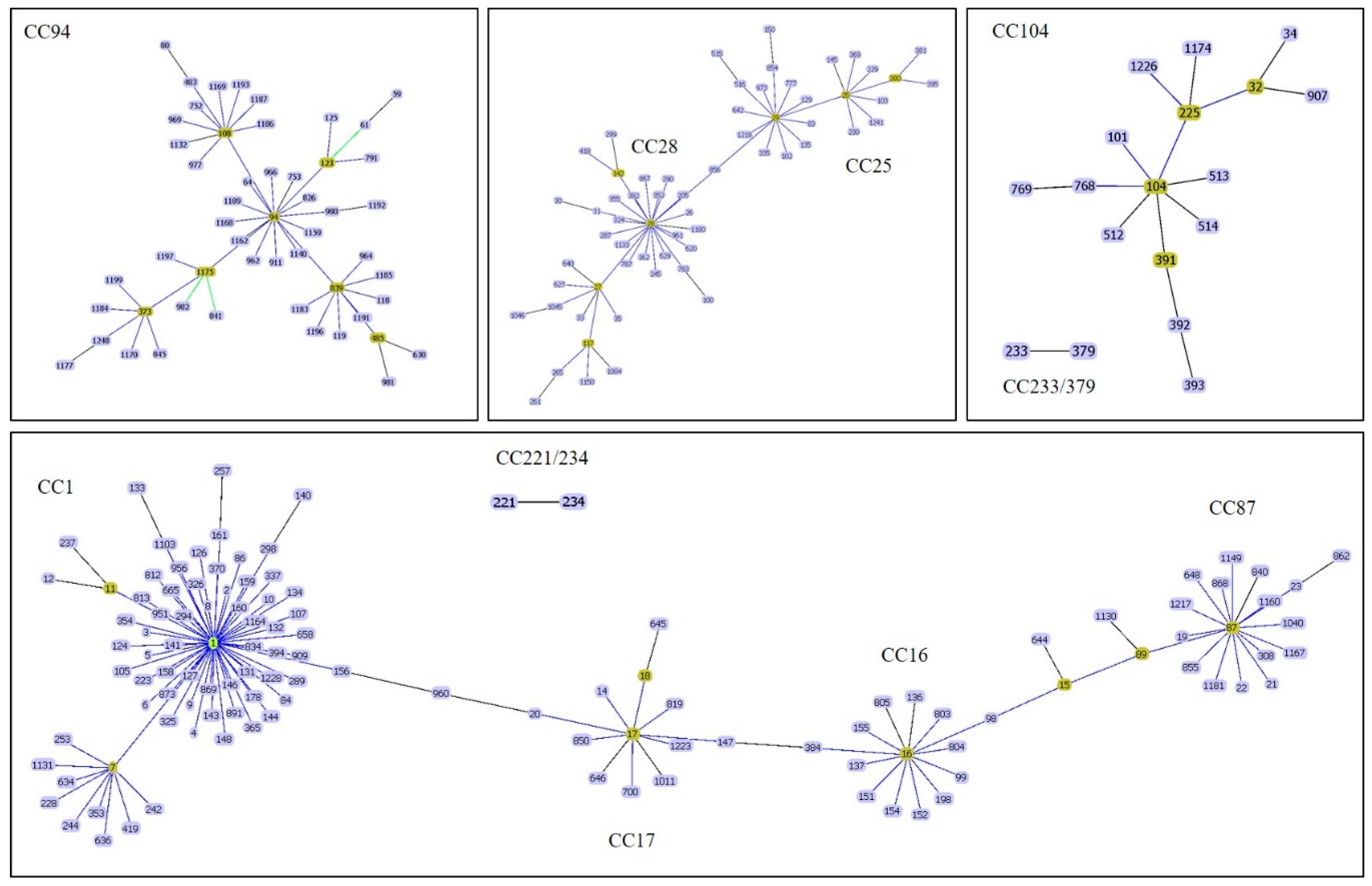
3. Prediction of CC Using Polymerase Chain Reaction
|
Clonal Complexes |
PCR Methods |
||||
|---|---|---|---|---|---|
|
Multiplex PCR |
PCR of ofs Genes |
PCR-Pilus-Associated Gene Profiles |
RAPD |
16S-23S rDNA PCR-RFLP |
|
|
CC1 |
Yes |
Yes |
Yes |
Yes |
Yes |
|
CC16 |
No |
No |
No |
Yes |
Yes |
|
CC20 ** |
No |
No |
No |
No |
No |
|
CC25 |
Yes |
Yes * |
Yes * |
Yes |
Yes * |
|
CC28 |
Yes |
Yes * |
Yes * |
Yes |
Yes * |
|
CC94 |
No |
Yes * |
No |
No |
No |
|
CC104 |
Yes |
Yes * |
Yes |
Yes |
Yes * |
|
CC233/379 |
Yes |
No |
No |
Yes |
Yes * |
|
CC221/234 |
Yes |
No |
No |
Yes |
Yes |
Note: * reveal the same profile; thus, could not be differentiated for each. ** PCR methods were not applied to CC20.
4. Whole-Genome Sequencing Approaches
References
- Goyette-Desjardins, G.; Auger, J.-P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes. Infect. 2014, 3, e45.
- Hlebowicz, M.; Jakubowski, P.; Smiatacz, T. Streptococcus suis Meningitis: Epidemiology, Clinical Presentation and Treatment. Vector Borne Zoonotic Dis. 2019, 19, 557–562.
- Huong, V.T.L.; Ha, N.; Huy, N.T.; Horby, P.; Nghia, H.D.T.; Thiem, V.D.; Zhu, X.; Hoa, N.T.; Hien, T.T.; Zamora, J.; et al. Epidemiology, Clinical manifestations, and outcomes of Streptococcus suis infection in Humans. Emerg. Infect. Dis. 2014, 20, 1105–1114.
- Dutkiewicz, J.; Sroka, J.; Zając, V.; Wasiński, B.; Cisak, E.; Sawczyn, A.; Kloc, A.; Wójcik-Fatla, A. Streptococcus suis: A re-emerging pathogen associated with occupational exposure to pigs or pork products. Part I—Epidemiology. Ann. Agric. Environ. Med. 2017, 24, 683–695.
- Gottschalk, M.; Higgins, R.; Jacques, M.; Mittal, K.R.; Henrichsen, J. Description of 14 new capsular types of Streptococcus suis. J. Clin. Microbiol. 1989, 27, 2633–2636.
- Gottschalk, M.; Higgins, R.; Jacques, M.; Beaudoin, M.; Henrichsen, J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J. Clin. Microbiol. 1991, 29, 2590–2594.
- Gottschalk, M.; Higgins, R.; Jacques, M.; Beaudoin, M.; Henrichsen, J. Isolation and characterization of Streptococcus suis capsular types 9–22. J. Vet. Diagn. Investig. 1991, 3, 60–65.
- Higgins, R.; Gottschalk, M.; Boudreau, M.; Lebrun, A.; Henrichsen, J. Description of Six New Capsular Types (29–34) of Streptococcus Suis. J. Vet. Diagn. Investig. 1995, 7, 405–406.
- Hill, J.E.; Gottschalk, M.; Brousseau, R.; Harel, J.; Hemmingsen, S.M.; Goh, S.H. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet. Microbiol. 2005, 107, 63–69.
- Tien, L.H.T.; Nishibori, T.; Nishitani, Y.; Nomoto, R.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA–DNA homology and sodA and recN phylogenies. Vet. Microbiol. 2013, 162, 842–849.
- Nomoto, R.; Maruyama, F.; Ishida, S.; Tohya, M.; Sekizaki, T.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22 and 26: Streptococcus parasuis sp. Int. J. Syst. Evol. Microbiol. 2015, 65, 438–443.
- Okura, M.; Osaki, M.; Nomoto, R.; Arai, S.; Osawa, R.; Sekizaki, T.; Takamatsu, D. Current Taxonomical Situation of Streptococcus suis. Pathogens 2016, 5, 45.
- Tohya, M.; Arai, S.; Tomida, J.; Watanabe, T.; Kawamura, Y.; Katsumi, M.; Ushimizu, M.; Ishida-Kuroki, K.; Yoshizumi, M.; Uzawa, Y.; et al. Defining the taxonomic status of Streptococcus suis serotype 33: The proposal for Streptococcus ruminantium sp. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 3660–3665.
- Hatrongjit, R.; Kerdsin, A.; Gottschalk, M.; Takeuchi, D.; Hamada, S.; Oishi, K.; Akeda, Y. First human case report of sepsis due to infection with Streptococcus suis serotype 31 in Thailand. BMC Infect. Dis. 2015, 15, 392.
- Kerdsin, A.; Oishi, K.; Sripakdee, S.; Boonkerd, N.; Polwichai, P.; Nakamura, S.; Uchida, R.; Sawanpanyalert, P.; Dejsirilert, S. Clonal dissemination of Streptococcus suis serotype 14 in Thailand. J. Med. Microbiol. 2009, 58, 1508–1513.
- Kerdsin, A.; Dejsirilert, S.; Sawanpanyalert, P.; Boonnark, A.; Noithachang, W.; Sriyakum, D.; Simkum, S.; Chokngam, S.; Gottschalk, M.; Akeda, Y.; et al. Sepsis and spontaneous bacterial peritonitis in Thailand. Lancet 2011, 378, 960. Available online: https://doi.org/10.1016/S0140-6736(11)60923-9 (accessed on 27 January 2020).
- Kerdsin, A.; Hatrongjit, R.; Gottschalk, M.; Takeuchi, D.; Hamada, S.; Akeda, Y.; Oishi, K. Emergence of Streptococcus suis serotype 9 infection in humans. J. Microbiol. Immunol. Infect. 2017, 50, 545–546.
- King, S.J.; Leigh, J.A.; Heath, P.J.; Luque, I.; Tarradas, C.; Dowson, C.G.; Whatmore, A.M. Development of a Multilocus Sequence Typing Scheme for the Pig Pathogen Streptococcus suis: Identification of Virulent Clones and Potential Capsular Serotype Exchange. J. Clin. Microbiol. 2002, 40, 3671–3680.
- Kerdsin, A.; Akeda, Y.; Takeuchi, D.; Dejsirilert, S.; Gottschalk, M.; Oishi, K. Genotypic diversity of Streptococcus suis strains isolated from humans in Thailand. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 917–925.
- Oh, S.-I.; Jeon, A.B.; Jung, B.-Y.; Byun, J.-W.; Gottschalk, M.; Kim, A.; Kim, J.W.; Kim, H.-Y. Capsular serotypes, virulence-associated genes and antimicrobial susceptibility of Streptococcus suis isolates from pigs in Korea. J. Vet. Med. Sci. 2017, 79, 780–787.
- Schultsz, C.; Jansen, E.; Keijzers, W.; Rothkamp, A.; Duim, B.; Wagenaar, J.A.; Van Der Ende, A. Differences in the Population Structure of Invasive Streptococcus suis Strains Isolated from Pigs and from Humans in the Netherlands. PLoS ONE 2012, 7, e33854.
- Groves, M.D.; Jordan, D.; Chapman, T.A.; Al Jassim, R. Multilocus sequence typing of Australian Streptococcus suis type 2 by MALDI-TOF mass spectrometry analysis of PCR amplicons. Vet. Microbiol. 2015, 177, 394–397.
- Oh, Y.; Tark, D.; Moon, S.-H.; Han, J.-I.; Kim, W.-I.; Cho, H.-S. Sepsis Caused by Streptococcus suis Serotype 2 in a Eurasian River Otter (Lutra lutra) in the Republic of Korea. J. Wildl. Dis. 2018, 54, 866–869.
- Estrada, A.A.; Gottschalk, M.; Rossow, S.; Rendahl, A.; Gebhart, C.; Marthaler, D.G. Serotype and Genotype (Multilocus Sequence Type) of Streptococcus suis Isolates from the United States Serve as Predictors of Pathotype. J. Clin. Microbiol. 2019, 57, e00377-19.
- Takamatsu, D.; Osaki, M.; Tharavichitkul, P.; Takai, S.; Sekizaki, T. Allelic variation and prevalence of serum opacity factor among the Streptococcus suis population. J. Med. Microbiol. 2008, 57, 488–494.
- Takamatsu, D.; Nishino, H.; Ishiji, T.; Ishii, J.; Osaki, M.; Fittipaldi, N.; Gottschalk, M.; Tharavichitkul, P.; Takai, S.; Sekizaki, T. Genetic organization and preferential distribution of putative pilus gene clusters in Streptococcus suis. Vet. Microbiol. 2009, 138, 132–139.
- Hatrongjit, R.; Kerdsin, A.; Gottschalk, M.; Hamada, S.; Oishi, K.; Akeda, Y. Development of a multiplex PCR assay to detect the major clonal complexes of Streptococcus suis relevant to human infection. J. Med. Microbiol. 2016, 65, 392–396.
- Maneerat, K.; Yongkiettrakul, S.; Kramomtong, I.; Tongtawe, P.; Tapchaisri, P.; Luangsuk, P.; Chaicumpa, W.; Gottschalk, M.; Srimanote, P. Virulence Genes and Genetic Diversity ofStreptococcus suisSerotype 2 Isolates from Thailand. Transbound. Emerg. Dis. 2013, 60, 69–79.
- Athey, T.B.T.; Teatero, S.; Lacouture, S.; Takamatsu, D.; Gottschalk, M.; Fittipaldi, N. Determining Streptococcus suis serotype from short-read whole-genome sequencing data. BMC Microbiol. 2016, 16, 162.
- Huang, W.; Wang, M.; Hao, H.; Yang, R.; Xie, J.; Su, J.; Lin, M.; Cui, Y.; Jiang, Y. Genomic epidemiological investigation of a Streptococcus suis outbreak in Guangxi, China, 2016. Infect. Genet. Evol. 2019, 68, 249–252.
- Du, P.; Zheng, H.; Zhou, J.; Lan, R.; Ye, C.; Jing, H.; Jin, D.; Cui, Z.; Bai, X.; Liang, J.; et al. Detection of Multiple Parallel Transmission Outbreak of Streptococcus suis Human Infection by Use of Genome Epidemiology, China, 2005. Emerg. Infect Dis. 2017, 23, 204–211.
- Willemse, N.; van der Ende, A.; Schultsz, C. Reinfection with Streptococcus suis analysed by whole genome sequencing. Zoonoses Public Health 2019, 66, 179–183.
- Athey, T.B.; Teatero, S.; Takamatsu, D.; Wasserscheid, J.; Dewar, K.; Gottschalk, M.; Fittipaldi, N. Population Structure and Antimicrobial Resistance Profiles of Streptococcus suis Serotype 2 Sequence Type 25 Strains. PLoS ONE 2016, 11, e0150908.
- Athey, T.B.T.; Auger, J.-P.; Teatero, S.; Dumesnil, A.; Takamatsu, D.; Wasserscheid, J.; Dewar, K.; Gottschalk, M.; Fittipaldi, N. Complex Population Structure and Virulence Differences among Serotype 2 Streptococcus suis Strains Belonging to Sequence Type 28. PLoS ONE 2015, 10, e0137760.
- Zheng, H.; Du, P.; Qiu, X.; Kerdsin, A.; Roy, D.; Bai, X.; Xu, J.; Vela, A.I.; Gottschalk, M. Genomic comparisons of Streptococcus suis serotype 9 strains recovered from diseased pigs in Spain and Canada. Vet. Res. 2018, 49, 1.
- Ruan, Z.; Feng, Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016, 44, D682–D687.
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361.
- De Greeff, A.; Wisselink, H.J.; De Bree, F.M.; Schultsz, C.; Baums, C.G.; Thi, H.N.; Stockhofe-Zurwieden, N.; Smith, H.E. Genetic diversity of Streptococcus suis isolates as determined by comparative genome hybridization. BMC Microbiol. 2011, 11, 161.
- Zheng, H.; Lan, R.; Zheng, X.; Cui, Z.; Liu, Z.; Bai, X.; Ji, S.; Gottschalk, M.; Xu, J. Comparative Genomic Hybridization Identifies Virulence Differences in Streptococcus suis. PLoS ONE 2014, 9, e87866.
- Weinert, L.A.; Chaudhuri, R.R.; Wang, J.; Peters, S.E.; Corander, J.; Jombart, T.; Baig, A.; Howell, K.J.; Vehkala, M.; Välimäki, N.; et al. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat. Commun. 2015, 6, 6740. Available online: https://doi.org/10.1038/ncomms7740 (accessed on 27 January 2020).
- Willemse, N.; Howell, K.J.; Weinert, L.A.; Heuvelink, A.; Pannekoek, Y.; Wagenaar, J.A.; Smith, H.E.; Van Der Ende, A.; Schultsz, C. An emerging zoonotic clone in the Netherlands provides clues to virulence and zoonotic potential of Streptococcus suis. Sci. Rep. 2016, 6, 28984.
