Dodder species (Cuscuta spp.) are holoparasites that have extensive material exchange with their host plants through vascular connections.
Parasitism represents a lifestyle in which parasitic plants obtain nutrients from hosts, causing serious biotic stresses and impacts on global agriculture. Cuscuta spp. (dodder) are rootless and leafless stem parasites throughout their lifecycle, and cannot survive independently due to their very limited or absent photosynthesis. Their wide host range includes vegetables, crops, and pastures, and they are malignant parasitic weeds. The dodder penetrates the host and forms a specific organ—the haustorium—for host attachment; the vascular connections established by the haustoria serve as an open hub for the exchange of various substances (e.g., water, nutrients, pathogens, systemic signals, and even macromolecules) between the two plants. This exchange is known as cross-species transmission.
1. Introduction
Given the importance of cross-species transmission for adaptation, interaction, and evolution in parasitic systems, the study of cross-species transmission has become a popular subject. Since the 1960s, researchers have performed many studies on cross-species transmission. For instance, viruses
[4,5][1][2] and phytoplasmas
[6][3] have long been known to be transferred between hosts and dodder. A large number of proteins have been shown to be transferred between hosts and dodder, and long-distance mobile proteins can even be transferred to the seeds of foreign plants and among dodder bridge-connected hosts
[7][4]. Systemic signals, including salt stress- and herbivory-induced signals, have also been reported to be transmitted from the dodder to the host plant, and even among dodder-connected hosts
[8,9,10][5][6][7]. In addition, recent studies on cross-species transmission at the transcription level have provided breakthrough insights into host–parasite interactions. The bidirectional mobility of large-scale mRNAs has been demonstrated between dodders and host plants, providing potential mechanisms for RNA-based interactions in symplastic connections
[11,12][8][9]. It has been shown that parasite microRNAs (miRNAs) can transfer into host plants and may act as virulence factors of host gene expression to promote the establishment of parasitic relationships
[13][10]. Small interfering RNAs (siRNAs) can also migrate into the parasite, where they decrease the expression of parasite genes, providing great potential for gene-editing-based dodder prevention
[14,15][11][12]. Despite the progress that has been made in detailing these processes, our understanding of the cross-species transmission and functional effects of non-coding RNAs (ncRNAs), such as long non-coding RNAs (lncRNAs), is still limited.
NcRNAs are a type of RNA that cannot encode proteins, but can still participate in various biological processes, such as cell growth, proliferation, differentiation, and apoptosis
[16,17,18,19][13][14][15][16]. These ncRNAs comprise regulatory and housekeeping ncRNAs, as well as ncRNAs of unknown function; the regulatory ncRNAs can be further sub-divided into several categories, including siRNAs, miRNAs, and lncRNAs, according to their size
[20,21][17][18]. In general, lncRNAs represent a large class of RNAs having transcripts longer than 200 nucleotides (nt) in length and poor protein-coding potential
[22,23][19][20]. Early studies questioned the importance of lncRNAs and regarded them as transcriptional “noise” but, at present, many thousands of lncRNAs—transcribed from locations throughout both plant and animal genomes—have been identified by tilling and RNA-seq analyses
[24,25,26][21][22][23]. These lncRNAs are classified into long intergenic non-coding RNAs (lincRNAs), intronic lncRNAs, sense, and antisense lncRNAs, according to their relative location with protein-coding genes
[27][24].
Regulatory roles for these lncRNAs in chromatin modification and transcription are currently under intense investigation
[24][21]. Studies have revealed that lncRNAs can coordinate gene expression, through a hormone–redox–cell wall network, to regulate growth process in plants, such as tomato fruit cracking
[28][25]. In
Arabidopsis thaliana (L.) Heynh.,
DROUGHT INDUCED lncRNA (DRIR) regulates the plant response to drought and salt stress as a novel positive regulator
[29][26]. LncRNAs can also participate in other abiotic stress responses in plants, such as heat stress, cold stress, and oxidative stress
[30,31,32,33][27][28][29][30]. A recent study has found that tomato
lncRNA23468 modulated the accumulation of
NBS-LRRs in the interaction between
Phytophthora infestans (Mont.) de Bary and tomato by decoying the expression of
miR482b, indicating that lncRNAs can also respond to biotic stresses
[34][31]. Although lncRNAs may play a broadly critical role in coordinating growth and development, as well as in abiotic and biotic responses, the biological significance of lncRNA movements remains largely elusive, with only a few studies having been carried out on the transport of lncRNAs. In plants, grafting studies have identified 22 lncRNAs which move systemically into root tips and developing leaves, where they can respond to early Pi deficiency
[35][32]. It has also been shown that lncRNAs are transferred between different types of cells through exosomes as a means of information exchange, acting as important activators or inhibitors to regulate gene expression and participating in a variety of biological processes
[36,37][33][34]. Thus, these observations that lncRNAs can move long distances through phloem to sink tissues, or move in different cells, have suggested to us the bold idea that lncRNAs might have potential mobility across species through dodder bridges, which merits further exploration.
Recently, the genomes of
C. australis R.Br. and
C. campestris Yunck. have been sequenced and published, thus providing useful resources for the comprehensive investigation of the evolution and physiological ecology of
Cuscuta [1,38][35][36]. Furthermore, the whole-genome sequence of crop soybean [
Glycine max (L.) Merr. var Williams 82], one of the known hosts of dodder, has been reported
[39,40][37][38]. This evidence provides support that soybean and dodder can be used as ideal candidate parasitic systems for further investigation of the ability of haustorium-mediated lncRNA transfer between two organisms.
21.1. Dodder Infestation-Induced Physiology Responses in Soybean Host
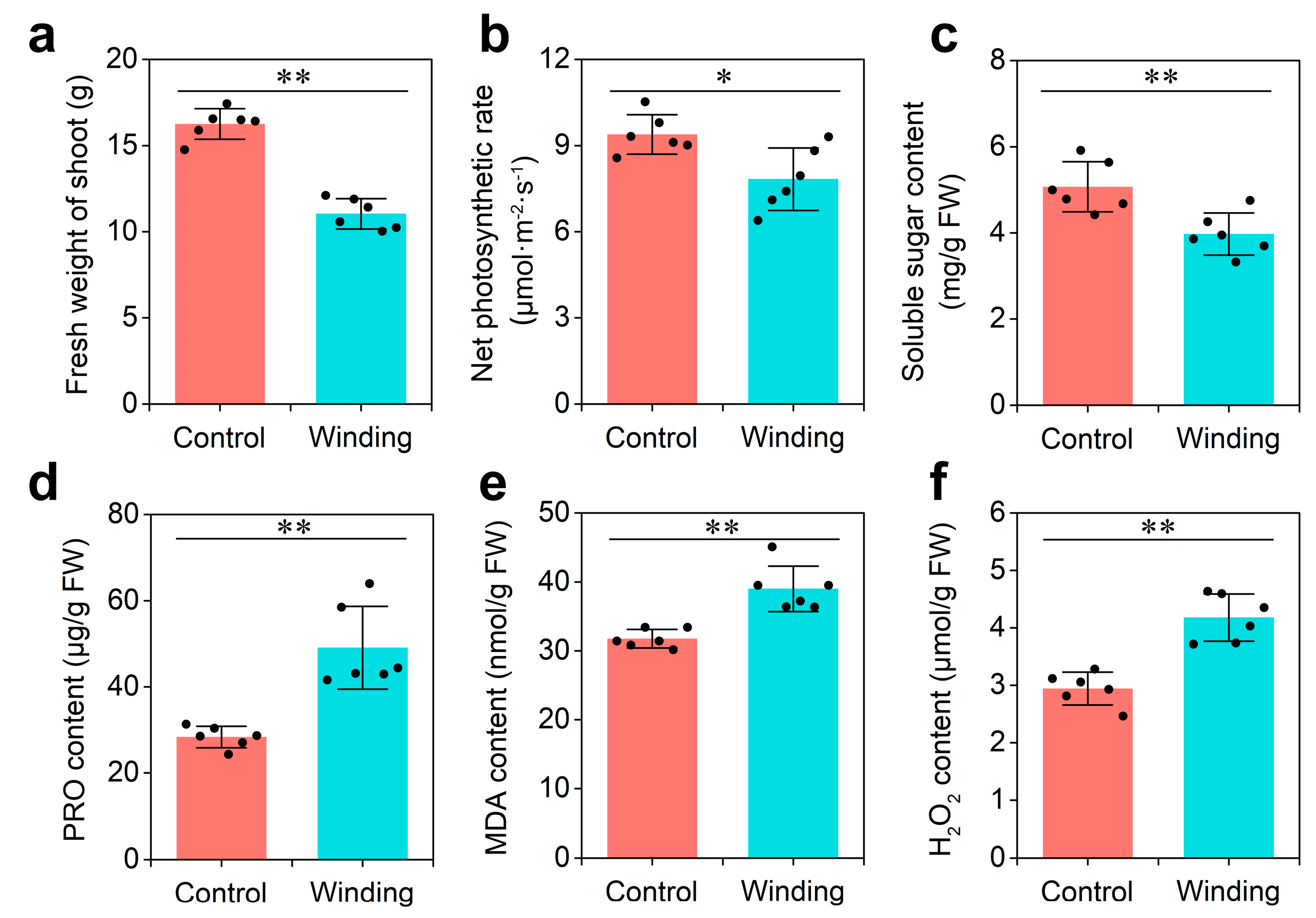
Dodder infestation has severe effects on the growth of its host. To explore the physiological responses of hosts to dodder parasitism, two-week-old soybean seedlings were infested with dodder (winding group) or mock-treated (control group) for 3 weeks. Compared with those in the control group, the fresh weight of shoots, net photosynthetic rate, and soluble sugar content of soybean infested by the dodder decreased significantly in the winding group (
Figure 1a–c). In contrast, proline (PRO), malondialdehyde (MDA), and H
2O
2 contents in the winding group were 75%, 22%, and 33% higher than in the control group, respectively (
Figure 1d–f). These data indicate that soybean plants prime themselves to respond dramatically to the dodder parasitism at the physiological level, which provides an important stepping stone in understanding lncRNA communication at the molecular level.
Figure 1. Physiological analysis of soybean in response to dodder parasitism: (a) Fresh weight of shoot; (b) net photosynthetic rate; (c) soluble sugar content; (d) proline (PRO) content; (e) malondialdehyde (MDA) content; and (f) H2O2 content. Asterisks indicate significant differences between control (soybean without dodder) and winding (soybean winded by dodder) groups, determined by Student’s t-test (n = 6; *, p < 0.05; **, p < 0.01). Error bars are ±SE.
2. RNA Sequencing and Identification of lncRNAs
In order to determine whether there exists cross-species lncRNA transfer in the soybean–dodder parasitization system, dodder seedlings were initially twisted and spread on soybean plants. Then, the dodder stems, interface stems where the parasite was connected to the soybean, and soybean stems were collected when the parasitic system had been established
(Supplementary Figure S1). Three biological repeats were performed for each group of samples, and nine samples were sequenced on an Illumina NovaSeq platform for transcriptome analysis. A total of 121.84 Gb of clean data were ultimately generated, after the removal of poor-quality reads and adapters. The clean sequences were used to identify lncRNAs present in the analyzed tissues. To this end, cleaned paired-end reads were mapped to the soybean reference genome (Wm82.a2.v1)
[40][38] and the
C. australis reference genome
[38][36]. Sequences that did not match any of the genomes due to sequencing errors were filtered out. Subsequently, reads that matched to both genomes and only matched to the native genome were considered to be from native transcripts, while reads that matched the foreign plant genome but not the native plant genome were considered to be mobile transcripts. After strict screening and mapping, the mapping rates were generally greater than 85%. These results indicated that the RNA-seq reads were highly reliable
(Supplementary Table S1).
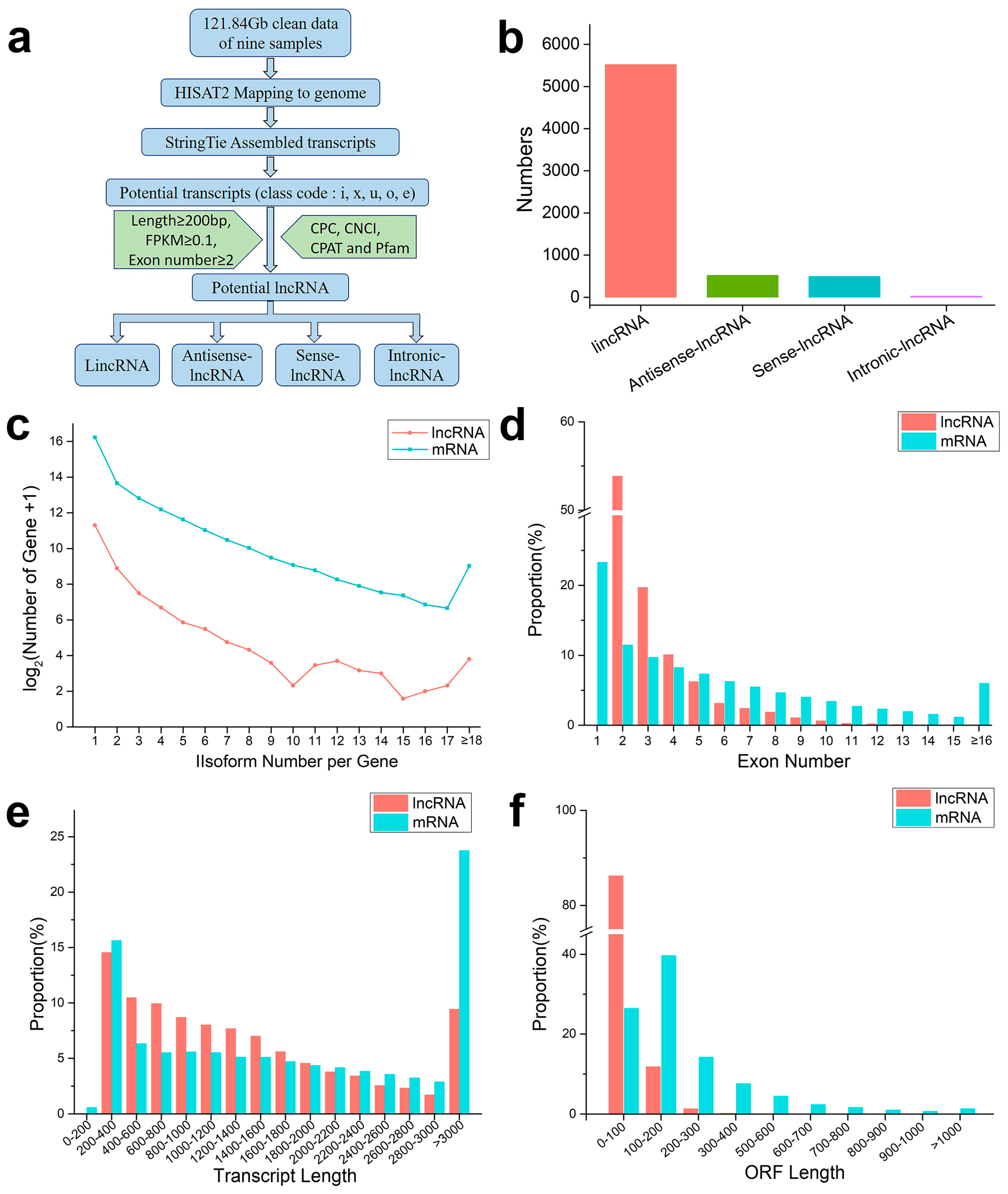
According to the pipeline in
Figure 2a, further analysis identified 6580 lncRNAs, including 1892 soybean lncRNAs and 4688 dodder lncRNAs. These lncRNAs were assigned to 5525 lincRNAs, 526 antisense lncRNAs, 497 sense lncRNAs, and 32 intronic lncRNAs, according to the anatomical properties of their gene loci (
Figure 2b). Subsequently, the basic genomic features of lncRNAs and mRNAs were comparatively analyzed. It was found that lncRNAs were expressed at similar levels in different groups and had fewer fragments per kilobase per million fragments mapped (FPKM) than protein-coding mRNAs in each group
(Supplementary Figure S2). Among them, 54% of the lncRNAs were spliced (
Figure 2c). The majority of lncRNAs (~55%) had two exons, and the number of lncRNAs decreased with an increase in the number of exons, while mRNAs contained more and more widely distributed exons: approximately 6% of mRNAs had more than 16 exons (
Figure 2d). The average length of these lncRNAs (1458 bp) was shorter than that of protein-coding mRNAs (2133 bp); approximately 60% of the lncRNA lengths ranged from 200 to 1400 bp, while those longer than 3000 bp accounted for only 9% (
Figure 2e). More than 90% of lncRNAs contained an open reading frame (ORF) of length ≤ 200 bp, while about of 34% mRNAs had ORF length ≥ 200 bp (
Figure 2f). Overall, both the transcript length and ORF length of lncRNAs were shorter, compared with those of mRNAs.
Figure 2. An integrative computational pipeline for the systematic identification and characterization of lncRNAs: (a) informatics pipeline for identification of lncRNAs; (b) composition of various types of lncRNAs; (c) number distributions of spliced lncRNAs and mRNAs; (d) proportion of exons per transcription for lncRNAs and mRNAs; (e) transcript length distributions for all lncRNAs and mRNAs; and (f) open reading frame (ORF) distributions for all lncRNAs and mRNAs.
3. Identification and Validation of Mobile lncRNAs
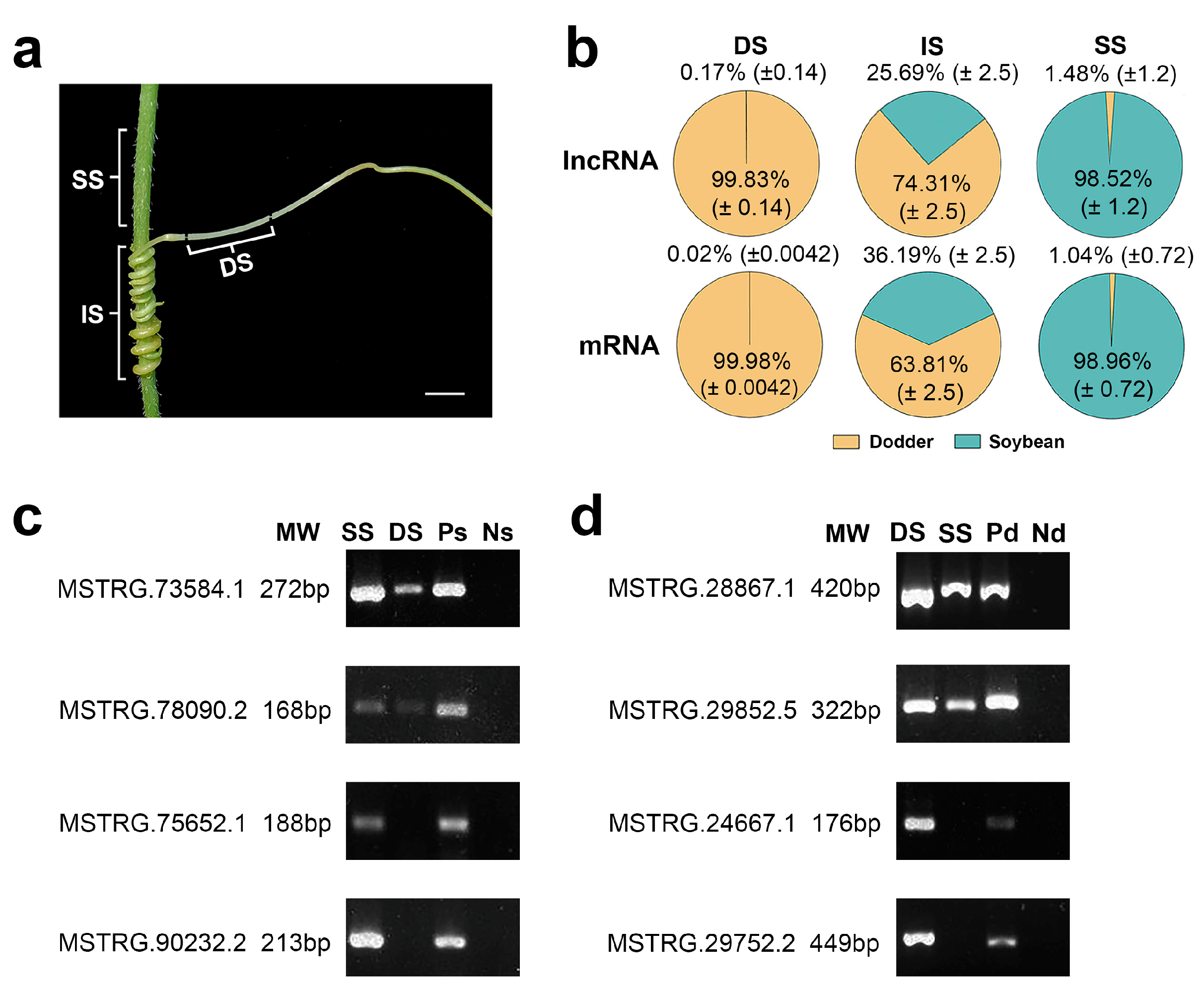
To explore the mobility of lncRNAs between the different plants, it used the above-developed lncRNA database to analyze the lncRNAs in three various tissues (dodder stems, soybean stems, and interface stems). In dodder stems, the proportions of the lncRNA reads from soybean averaged 0.17% of the total mapped reads across three sequencing runs, whereas soybean stems contained 1.48% dodder lncRNA reads, indicating that bidirectional movement of lncRNAs occurred between the dodder and soybean. Similarly, dodder stems contained 0.02% soybean mRNA reads, while soybean stems contained 1.04% dodder mRNA reads, suggesting that lncRNA movement is usually accompanied by mRNA trafficking (
Figure 3a,b
; Supplementary Table S2).
Figure 3. Transcript transfer in soybean–dodder parasitization systems: (a) Sequencing and analysis of three types of tissues, including the dodder stems (DS), interface stems (IS), and soybean stems (SS). Scale bars = 5 mm; (b) Pie charts illustrating the proportion of reads from foreign and native lncRNAs or mRNAs in each tissue. Calculations were based on the ratio of the reads mapped only to the foreign genome and total reads mapped to the foreign and native genomes. The values are the mean ± standard deviation of three replicates. DS, IS, and SS represent dodder stems, interface stems, and soybean stems, respectively; (c) RT-PCR confirmed the transfer of lncRNAs into the dodder for two soybean lncRNAs, MSTRG.73584.1 and MSTRG.78090.2; MSTRG.75652.1 and MSTRG.90232.2 were not detected in the dodder by RNA-seq. SS represents soybean stem; DS represents dodder stem; Ps represents positive control (soybean without dodder); Ns represents negative control (dodder not growing on soybean); (d) RT-PCR confirmed the transfer of lncRNAs into the host for two dodder lncRNAs, MSTRG.28867.1 and MSTRG.29852.5; MSTRG.24667.1 and MSTRG.29752.2 were not detected in soybean by RNA-seq. DS represents dodder stem; SS represents soybean stem; Pd represents positive control (dodder not growing on soybean); Nd represents negative control (soybean without dodder).
In the dodder–soybean parasitic system, the established mobile reads represent the diversity of transcripts. Subsequently, the number of mobile or non-mobile transcripts was determined, in order to compare the transferability of inter-plant lncRNAs and mRNAs. As shown in
Table 1, 365 dodder lncRNAs and 8894 dodder mRNAs were detected in soybean stems, accounting for 7.8% (365/4688) and 52.4% (8894/16,977) of the total dodder lncRNAs and mRNAs, respectively. In contrast, only 14 soybean lncRNAs and 74 soybean mRNAs were identified in dodder stems, comprising 0.74% (14/1892) and 0.17% (74/42,296) of the total transcripts of soybean, respectively.
Table 1. Numbers of lncRNAs and mRNAs transferred in the soybean–dodder system.
| Mobility Category |
Soybean lncRNAs |
Dodder lncRNAs |
Soybean mRNAs |
Dodder mRNAs |
| Total mobile |
14 |
365 |
74 |
8894 |
| Nonmobile |
1878 |
4323 |
42,222 |
8083 |
| Total |
1892 |
4688 |
42,296 |
16,977 |
To further confirm the trafficking of inter-plant lncRNA individuals, several mobile and non-mobile lncRNA transcripts were selected and analyzed by reverse transcription-polymerase chain reaction (RT-PCR). Mobile lncRNAs MSTRG.73584.1 and MSTRG.78090.2 from soybean were detected in dodder stems at a lower level than in soybean stems; similarly, mobile lncRNAs MSTRG.28867.1 and MSTRG.29852.5 from dodder were detected in soybean stems at a lower level than in dodder stems. In contrast, non-mobile lncRNAs were detected only in soybean stems or dodder stems (
Figure 3c,d). The RT-PCR results indicated that the lncRNA data obtained by RNA-seq were reliable.
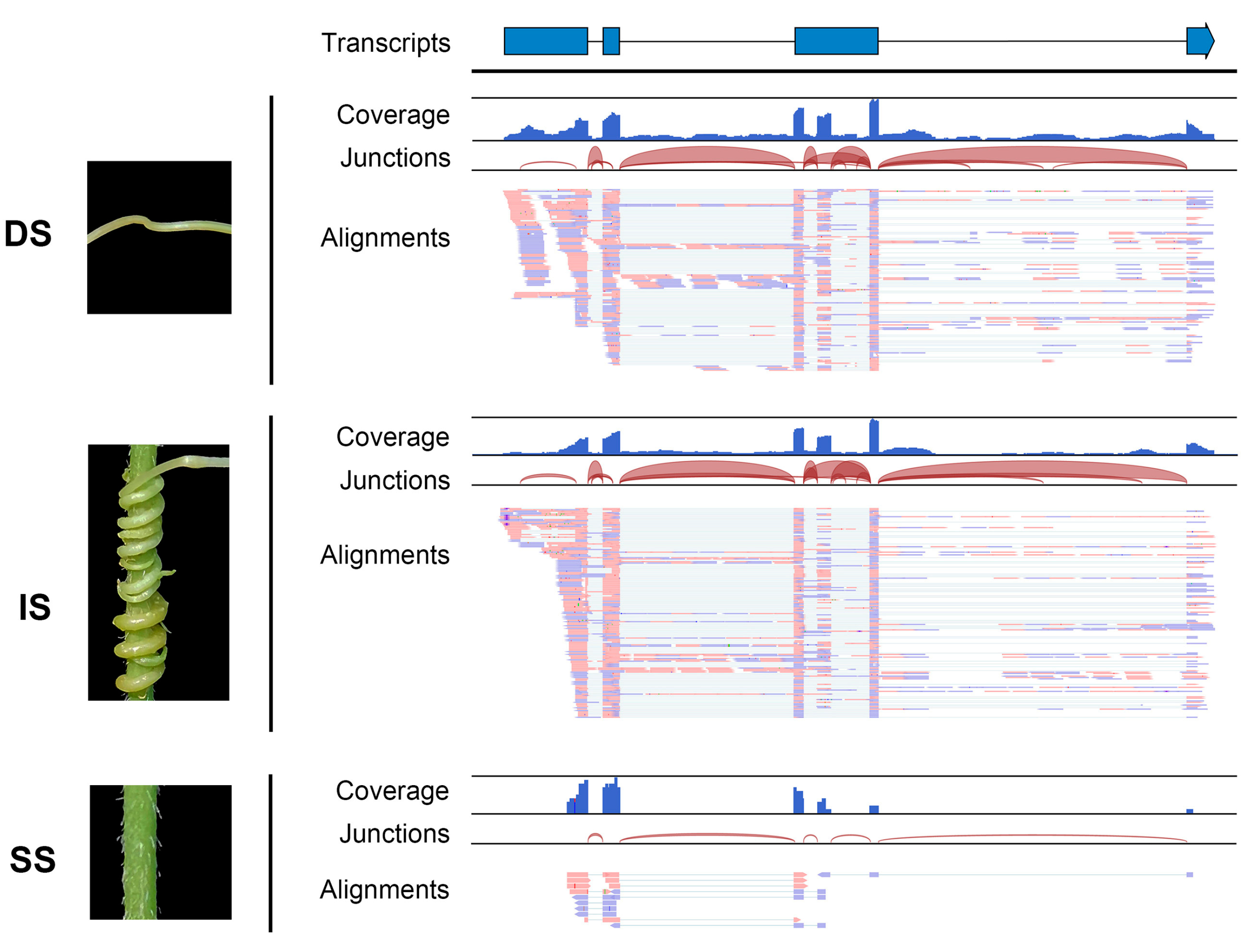
Additionally, the read coverage and alignments of RNA-seq data illustrated the form of the mobile transcripts. The read sequences and coverage of mobile lncRNA MSTRG.10219.19 from dodder stem tissue closely matched those of the interface tissue, with the exception that the mobile lncRNAs in the soybean stem tissue appeared in a fully spliced mature form; introns were only found in the libraries of dodder stems or interface tissues (
Figure 4). This further confirmed the actual movement of lncRNAs between the dodder and soybean. Notably, although the output of read mapping itself produced an attractive picture of lncRNA movement, such confirmation is not practical for all mobile lncRNAs.
Figure 4. Visualization of read assemblies of the dodder lncRNA MSTRG.10219.19 in three tissues. The lncRNA model at the top indicates exons as blue bars and introns as line bridges. Each panel includes tracks for total coverage, junction coverage, and read alignments. Reads that span junctions are connected with thin lines. DS, IS, and SS represent dodder stems, interface stems, and soybean stems, respectively.
4. General Properties of the Mobile Transcripts
Next, it investigated whether the inter-plant mobile transcripts possess certain properties that enable them to be transferred. First of all, by comparing the expression abundance of mobile and non-mobile transcripts in the interface stems, it was found that the abundance of mobile lncRNAs was higher than that of non-mobile lncRNAs in the interface stems, and the expression patterns of mRNAs were similar to those of lncRNAs (
Figure 5a,b).
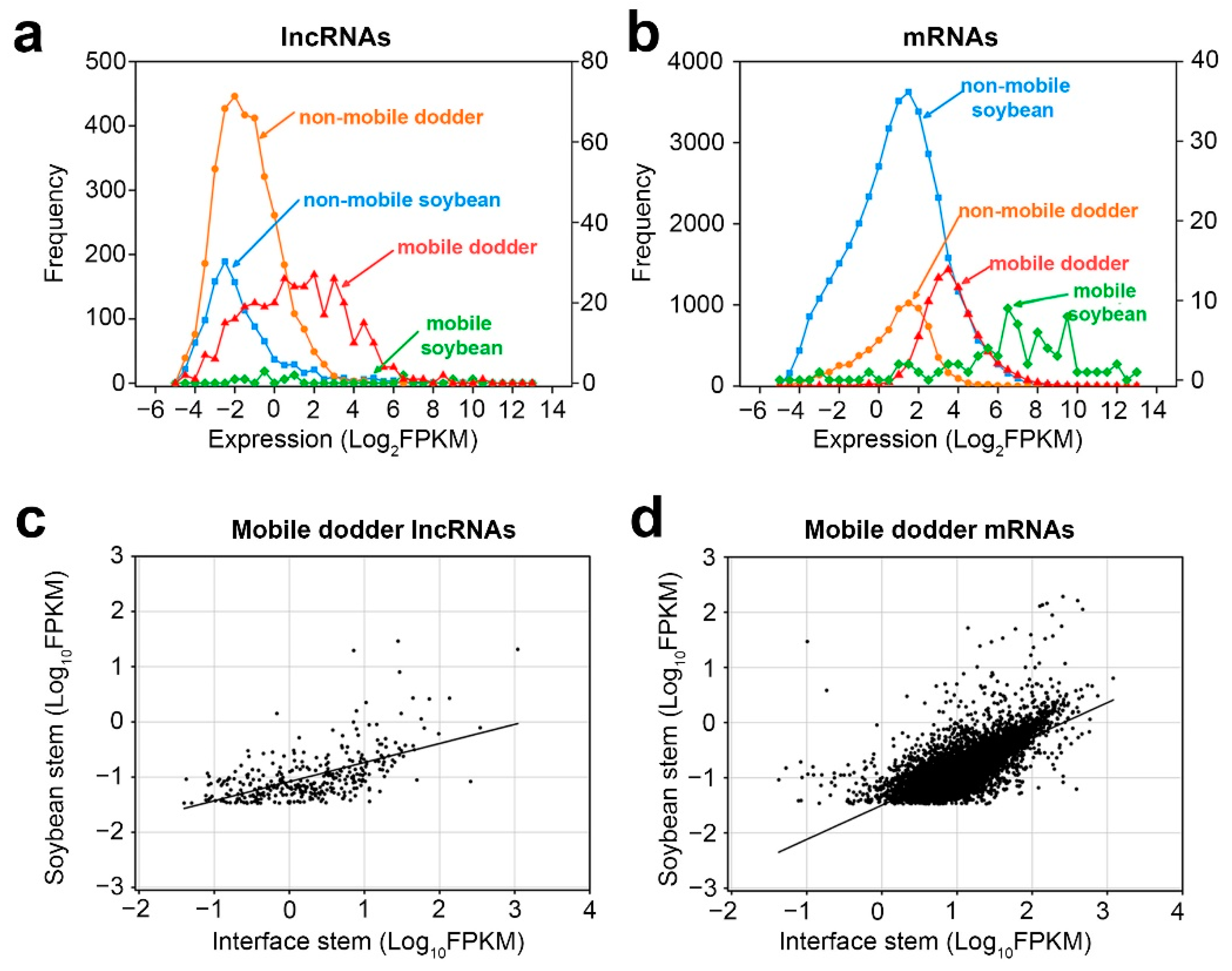
Figure 5. Properties of mobile and non-mobile transcripts: (a,b) distribution of lncRNAs (a) and mRNAs (b) transcript levels in interface stems related to mobility in dodder–soybean associations; (c,d) Scatter plots of lncRNAs (c) and mRNAs (d) transcript levels in the soybean stem versus those in interface stems. A total of 365 dodder lncRNAs were transferred into soybean, whereas 8894 dodder mRNAs were transferred into soybean. Lines correspond to linear regression analysis of the data.
Secondly, as there were only a small number of mobile soybean transcripts, correlation analysis was only performed for the transcript levels of mobile dodder lncRNAs and mRNAs in interface stems and soybean stems, respectively (
Figure 5c,d). The results showed that the expression levels of mobile dodder lncRNAs or mRNAs in interface stems had a positive linear correlation with those in soybean stems. Nonetheless, the mobility pattern of lncRNAs was more dispersed, whereas the mobility pattern of mRNAs was more focused around the regression line, indicating that the dynamics of transmission of lncRNAs may differ from those of mRNAs (
Figure 5c,d).
5. Functional Prediction of Mobile lncRNAs by Their Target Genes
To investigate the potential systemic roles of transfer lncRNAs, the target genes of transfer lncRNAs were predicted. LncRNAs spaced near protein-coding genes could participate in transcriptional regulation by binding to promoters and other
cis-acting elements
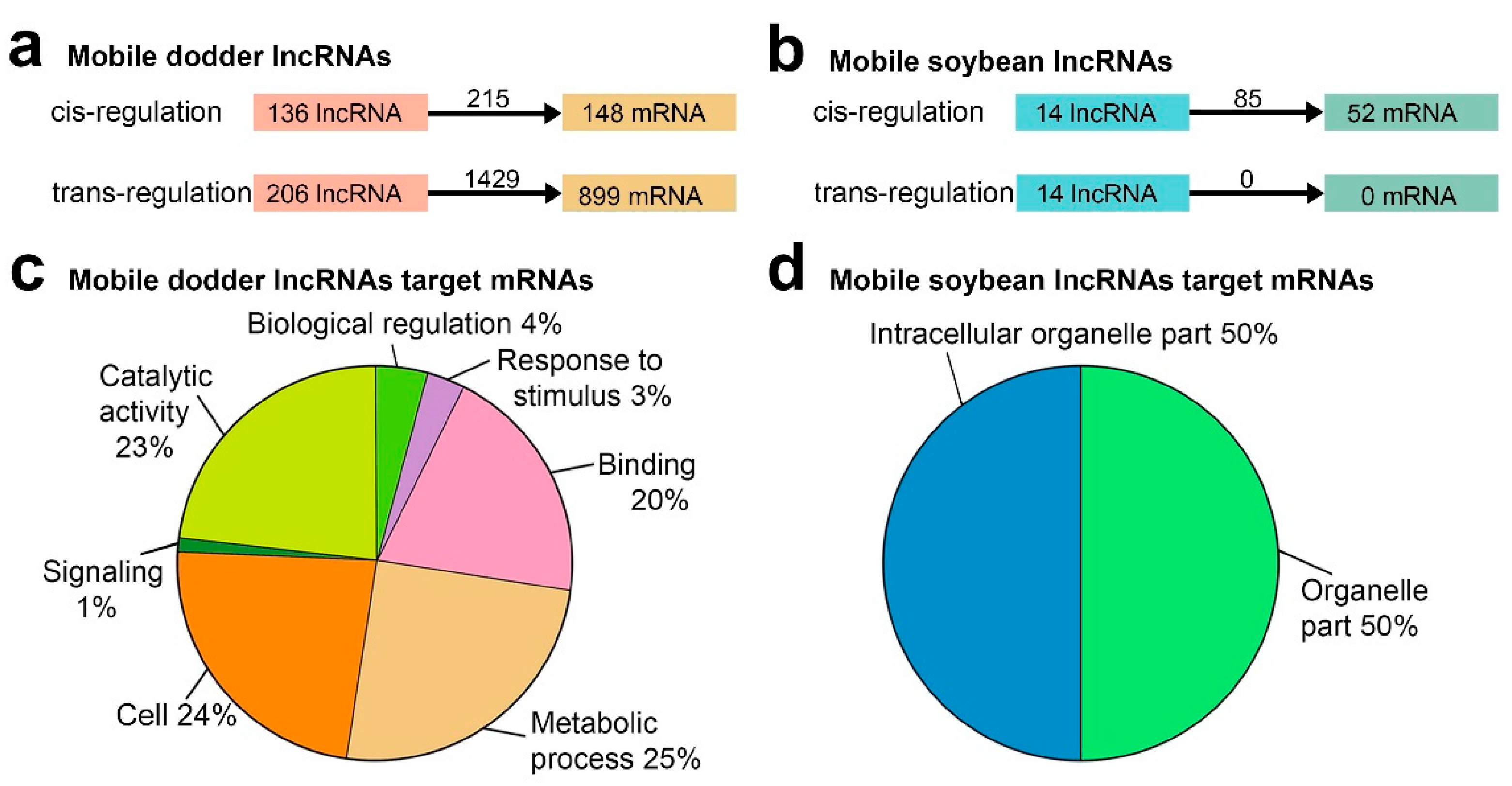
[27][24]. Thus, it first searched for the upstream and downstream 100 kb regions of lncRNAs and found that 136 mobile dodder lncRNAs might regulate 148 mRNAs with 215 lncRNA–mRNA pairs in
cis, and that 14 mobile soybean lncRNAs might regulate 52 mRNAs with 85 lncRNA–mRNA pairs in
cis, respectively (
Figure 6a,b
; Supplementary Table S3). Recently published data has suggested the great potential of detecting lncRNA-mediated regulation by base pair complementarity
[41][39], which was determined to identify
trans-acting lncRNAs. In total, 206 mobile dodder lncRNAs might regulate 899 mRNAs with 1429 lncRNA–mRNA pairs in
trans (
Figure 6a,b
; Supplementary Table S3). Furthermore, the expression patterns of mobile lncRNA target genes associated with three different tissues (dodder stems, interface stems, and soybean stems) were further analyzed using the MultiExperiment Viewer 4.9 (MEV 4.9) software
(Supplementary Figure S3). In addition, a total of 440 dodder target genes, including 70 (47.3%)
cis-target genes and 370 (41.2%)
trans-target genes, were predicted to be co-transferred with 159 mobile lncRNAs from dodder into soybean, while only four soybean
cis-target genes were predicted to be co-transferred with 11 mobile lncRNAs from soybean into dodder
(Supplementary Figure S4; Supplementary Table S3).
Figure 6. Functional analysis of mobile lncRNAs in parasitic systems: (
a) Schematic diagram of mobile dodder lncRNAs regulating mRNAs; (
b) Schematic diagram of mobile soybean lncRNAs regulating mRNAs. The numbers of regulatory relationship pairs are shown on the black arrows; (
c) Pie charts showing the percentages of Gene Ontology (GO) slim terms enriched by mobile dodder lncRNAs target genes by WEGO 2.0 (
p-value < 0.05); (
d) Pie charts showing the percentages of GO slim terms enriched by mobile soybean lncRNAs target genes by WEGO 2.0 (
p-value < 0.05).
The full list of GO slim terms for these data is presented in Supplementary Table S4a.
In order to gain insight into the function of these mobile lncRNAs, it then applied Gene Ontology (GO) enrichment to analyze their predicted target genes. A total of 12 mobile dodder and two mobile soybean GO terms were enriched by WEGO 2.0 (
p-value < 0.05
; Supplementary Table S4a). Notably, the great majority of the target genes of mobile dodder lncRNAs were enriched in “metabolic process”, “catalytic activity”, “signaling”, and “response to stimulus” categories, whereas the genes corresponding to mRNAs targeted by mobile soybean lncRNAs were only enriched in organelle-related categories, including “intracellular organelle part” and “organelle part” (
Figure 6c,d). In addition, the GO enrichment analysis of these target mRNAs was also performed using the agriGO 2.0 website, the results of which were similar to those found in WEGO enrichment analysis
(Supplementary Figure S5).
We also applied GO enrichment to assess the functional significance of mobile mRNAs using the WEGO 2.0 and agriGO 2.0 websites (Supplementary Table S4b). Similarly, most of the mobile dodder mRNAs were enriched in “metabolic process”, “catalytic activity”, “binding”, “biological regulation”, “response to stimulus”, and “signaling” categories (Supplementary Figure S6a; Supplementary Figure S7), while the mobile soybean mRNAs were similar to the corresponding lncRNAs, with some of them being enriched in “organelle” and “binding” categories (Supplementary Figure S6b; Supplementary Figure S7).
6. Identification of Transcription Factors of the Mobile Transcripts
Transcription factors (TFs) are regulatory proteins that can activate or inhibit target genes and which participate in biotic or abiotic stress responses
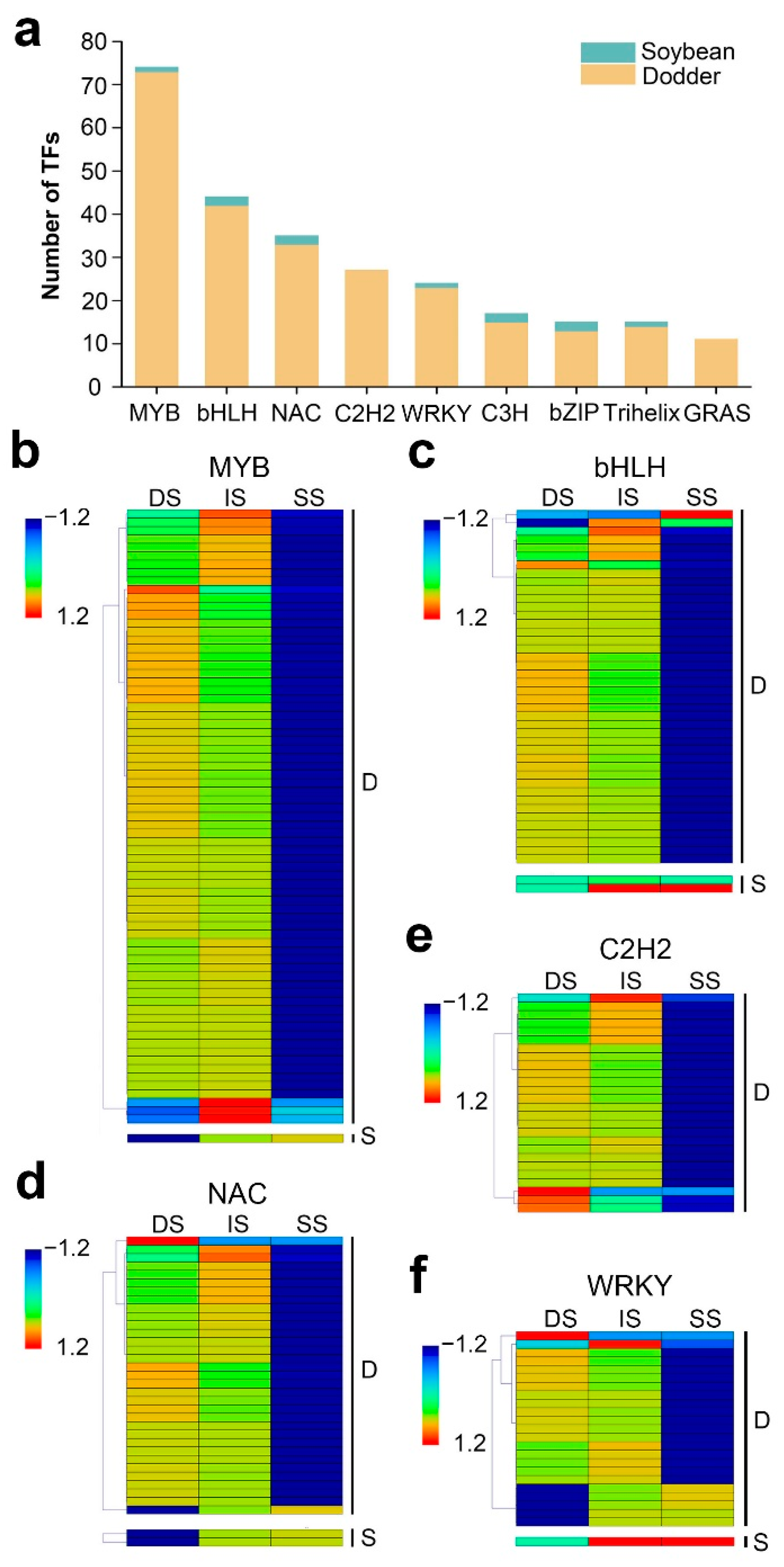
[42,43,44,45][40][41][42][43]. To further reveal the potential regulation functions of lncRNAs, it screened the TFs corresponding to their target mRNAs. In total, 201 mobile lncRNAs resulted in the identification of 635 targeted TFs, belonging to 49 TF families
(Supplementary Table S5a). In the dodder–soybean parasitic system, the MYB family was the largest gene family identified (74 in total), corresponding to 34 mobile lncRNAs, followed by the bHLH, NAC, C2H2, and WRKY families and presenting a high number of mobile transcripts (
Figure 7a). The dynamic changes in the expression levels of these TFs in the three different tissues are shown in
Figure 7b–f. In addition, when it screened the TFs for the mobile mRNAs, 54 TF families, including 297 mobile mRNAs, were shown to be transferred from dodder to soybean, while no TFs were predicted to be transferred from soybean to dodder
(Supplementary Table S5b). It was found a total of 30 TF families that were common to mobile lncRNA-targeted mRNAs and mobile mRNAs
(Supplementary Table S5).
Figure 7. Distribution and expression patterns of transcription factors: (a) Details of the number for TFs identified from mobile lncRNAs target genes of dodder and soybean. Data are sorted by number of lncRNAs. Only categories with more than 10 mobile transcripts identified as transcription factors are shown; (b–f) Heatmap of the expression patterns of the first five most numbers of TFs in the tissues of dodder stems (DS), interface stems (IS), and soybean stems (SS), including MYB (b), bHLH (c), NAC (d), C2H2 (e), and WRKY (f). ‘D’ represents mobile dodder lncRNAs target genes. ‘S’ represents mobile soybean lncRNAs target genes. The gene expression is based on the z-scores of log2(FPKM) value. The blue and red colors indicate low and high expression levels, respectively.
7. Potential lncRNA–mRNA/TF Network in Parasitic System
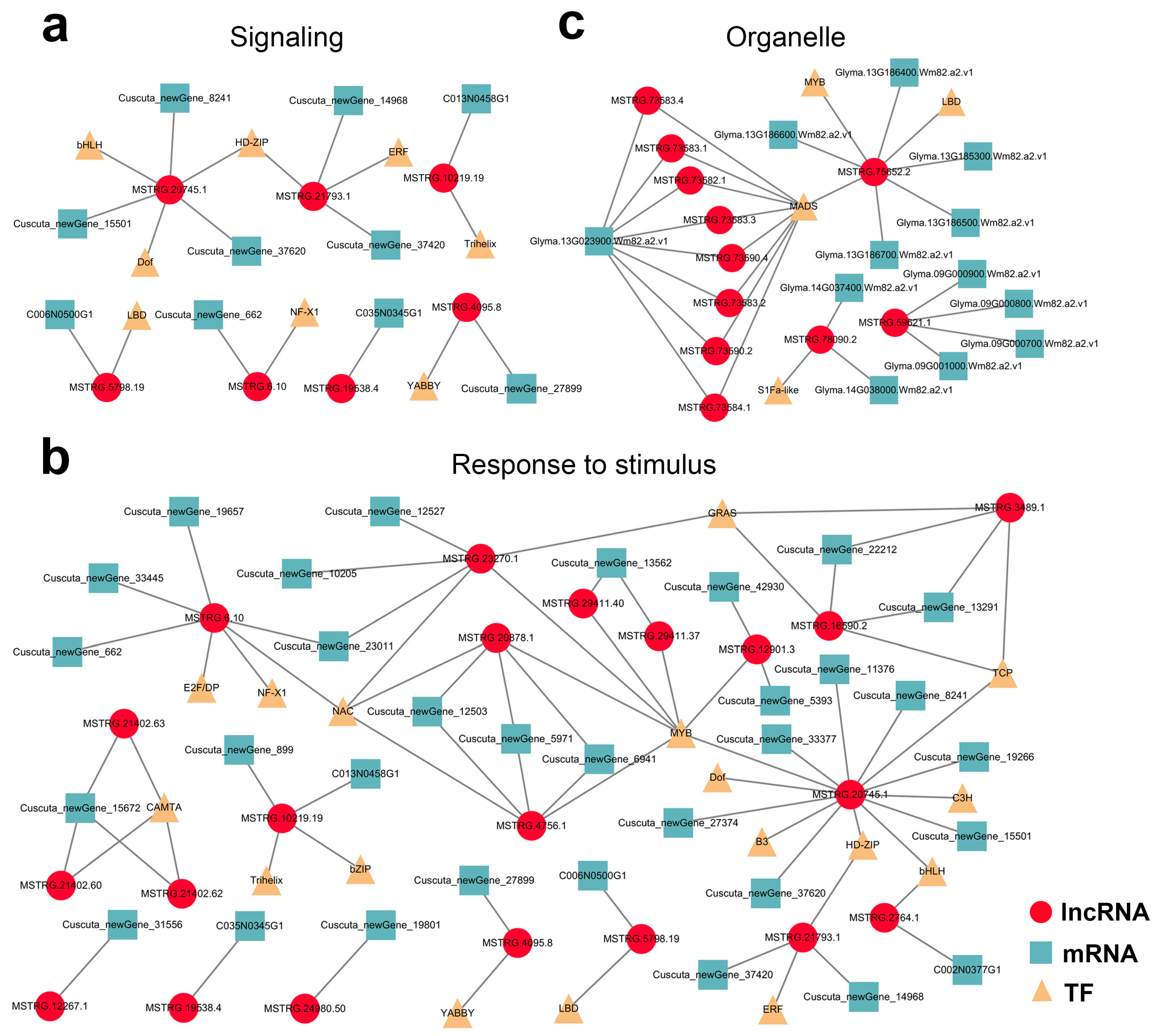
Finally, mobile lncRNA-associated networks were constructed based on all the target genes, revealing the intricacy of their relationships
(Supplementary Figure S8). Among them, the detailed interactions of the mobile dodder lncRNAs enriched in “signaling” and “respond to stimulus” terms (
Figure 8a,b), and the interactions of the mobile soybean lncRNAs enriched in organelle-related terms were visualized (
Figure 8c). In these networks, lncRNA target prediction revealed the presence of 5 and 24 potential lncRNA–mRNA target pairs that co-transferred from dodder to soybean in “signaling” and “respond to stimulus” terms, respectively. In contrast, there were 19 potential lncRNA–mRNA/IF target pairs enriched in organelle-related terms, but no lncRNA–mRNA/TF target pairs appeared to be co-transferred.
Figure 8. Potential lncRNA–mRNA/TF network in parasitic system visualized using Cytoscape 3.7.2: (a,b) Predicted network of mobile dodder lncRNAs and their targeted mRNAs/TFs enriched in “signaling” term (a) or “respond to stimulus” term (b); (c) Predicted network of mobile soybean lncRNAs and their targeted mRNAs/TFs enriched in “organelle part” term. Red circles represent lncRNAs, blue squares represent mRNAs, and yellow triangles represent TFs.