The pseudo-rotatis entry summarizes the synthetic efforts on aconal flexibility of the ribonucleotide is considerably limited due to the anomeric effect, and RNA/RNA and RNA/DNA duplexes are generally more thermally stable than DNA/DNA duplexes. The rigidity of the cyclic scaffold has been considered important for the formation of thermally stable duplexes, and the unexpectedly high thermal stability of duplexes formed with the participation of LNA oligomers could serve as an excellent justification for this point of view. However, this generalization is not consistent with the behavior of Peptide Nucleic Acids (PNA), in which the heterocyclic bases are attached to a linear peptide-like backbone, since duplexes composed of RNA or DNA and PNA strands are far more stable than RNA/RNA and DNA/DNA ones. This phenomenon may be attributed to the absence of a negative charge in the backbone, such that the absence of repulsive interactions balances the entropic cost of proper spatial organization of the flexible PNA scaffolds. Nonetheless, the widely accepted importance of the cyclic sugar components for the stability of the duplexes could be questioned. There is another perspective that can be applied to the acyclic analogs of nucleic acids and that is related to the origin of life. The synthetic efforts on acyclic analogs of nucleic acids and provides information on the most interesting features of selected classes of such compounds, are here described. The selection includes the following types of analogs: Flexible (FNA), Unlocked (UNA), Glycol (GNA), Butyl (BuNA), Threoninol (TNA) and Serinol Nucleic Acids (SNA). These classes of analogs are discussed in terms of their synthetic methods, the thermal stability of their homo- and hetero-duplexes and their applicability in biological and biochemical research and nanotechnology.
- acyclic nucleic acids
- stability of nucleic acid complexes
- fluorescent nucleic acid probes
- siRNA
Acyclic nucleic acids with phosphodiester linkages.
Agnieszka Tomaszewska-Antczak* and Piotr Guga
Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences, Sienkiewicza 112, 90-363 Lodz, Poland;
atom@cbmm.lodz.pl, pguga@cbmm.lodz.pl
* Correspondence: atom@cbmm.lodz.pl; Tel.: +48-(42)-6803215
Keyw1. Intrords: acyclic nucleic acids; stability of nucleic acid complexes; fluorescent nucleic acid probes; siRNA
uction
- Introduction
Nucleotide units present in DNA and RNA consists of three segments that are chemically very different from each other. The heterocyclic nucleobases serve as acceptors and donors of hydrogen bonds and are responsible for proper intermolecular recognition. In addition, stacking interactions are important in conferring a high degree of rigidity to oligomers and higher -order structures. The anionic phosphodiester groups are reasonably stable under mild hydrolytic conditions, but they are important for hydration and may be involved in electrostatic interactions with positively charged amino acid side chains and other cationic species. Not to be neglected is the electrostatic repulsion between the phosphate moieties present in the oligonucleotide chains forming a complex, which contribute to the rigidity of the entire DNA or RNA system. The third component comprises of sugar moieties (ribose or 2’-deoxyribose), which are present in the cyclic form. Notably, the pseudo-rotational flexibility of the ribonucleotide is considerably limited due to the anomeric effect, and RNA/RNA and RNA/DNA duplexes are generally more thermally stable than DNA/DNA duplexes. The rigidity of the cyclic scaffold has been considered important for the formation of thermally stable duplexes [1]. However, this generalization is not consistent with the behavior of Peptide Nucleic Acids (PNA), in which the heterocyclic bases are attached to a linear peptide-like backbone, since duplexes composed of RNA or DNA and PNA strands are far more stable than RNA/RNA and DNA/DNA ones. This phenomenon may be attributed to the absence of a negative charge in the backbone, such that the absence of repulsive interactions balances the entropic cost of proper spatial organization of the flexible PNA scaffolds. Nonetheless, the widely accepted importance of the cyclic sugar components for the stability of the duplexes could be questioned. There is another perspective that can be applied to the acyclic analogs of nucleic acids that is related to the origin of life. We are currently witnessing an ongoing dispute about which molecule, DNA or RNA, was the first to mark the beginning of life in its modern form, i.e., were the first carriers of the contemporary genetic information. However, even if the correct answer is finally found, there is still the question of its possible predecessors. The first suggestion that oligomers with an acyclic flexible scaffold carrying pro-chiral nucleotide analogs might play such a role was made in 1987 by Joyce et al. [2]. These flexible structures seemed to be easier to produce than deoxyribo- or ribonucleosides under prebiotic conditions. Thus, both perspectives triggered efforts to synthesize and evaluate numerous acyclic analogs of nucleic acids, and it must be admitted that in several cases, very astonishing properties were found.
- Flexible Nucleic Acids (FNA)
2. Flexible Nucleic Acids (FNA)
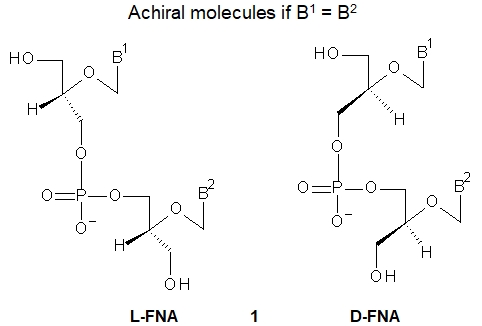
Flexible Nucleic Acids (FNA, 1, shown are dinucleotides) were one of the first proposed acyclic nucleotides for an evolutionary precursor to DNA and RNA, and were the first synthesized acyclic analogs of DNA. While glycerol itself is an achiral molecule, any derivatization at either of the two primary hydroxyl groups leads to a chiral compound producing L-FNA or D-FNA depending on the chiral glycerol derivative used in the synthesis.
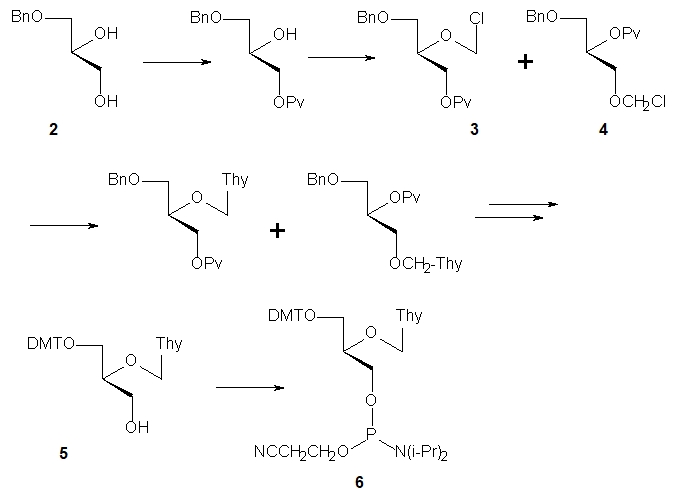
Scheme 1. Synthesis of the FNA-T phosphoramidite monomer.
Schneider and Benner [3] used (S)-1-O-benzyl glycerol (2, Scheme 1) to prepare the phosphoramidite monomer 6, which was used (with the approximately 90% coupling yields) in the solid phase synthesis of oligomers of the general sequences 5’-d(CTTTTTTTG)-3’ and 5’-d(CAAATAAAG)-3’. These oligomers consisted mainly of DNA, with the FNA unit at position 4 or two FNA units at positions 4,6 or 4,5. A 13-mer 5’-d(CtttttttttG)-3’ (“t” stands for the FNA-T unit) was also obtained. It was found that the oligomers containing 1 or 2 FNA-T units destabilized the (FNA-DNA)/DNA duplexes by about 15 °C per modification. The almost completely modified oligomer d(CtttttttttG) did not form a duplex with the complementary 5’-d( GAAAAAAAAAAAC)-3’. Destabilization was attributed to an increase in entropy cost compared to the perfect duplex formed by natural nucleic acids. Pure oligothymidylate and oligoadenylate FNA and their alternating copolymers (12–19 mers) were synthesized by Merle and collaborators [4]. Thermal dissociation studies showed that (FNA-A)12 hybridized effectively with dodecathymidylate (dT)12 and the complex exhibited sharp melting with Tm = 24 °C. Rather unexpectedly, although DNA/DNA duplexes adopt a B-type conformation, the circular dichroism (CD) spectrum suggested that the FNA/DNA duplex exists as an A-type helix.
Chaput and Switzer studied the nonenzymatic oligomerization reaction on hairpin templates containing several FNA-C units, which was considered a model for studying prebiotic replication of informational macromolecules [5]. It was found that the acyclic FNA-C units effectively attracted guanosine 5’-phosphoro-2-methylimidazolide molecules and did not block the template-directed oligomerization. PAGE Analysis performed after 10 days showed elongation by up to 7 nucleotides.
- Unlocked Nucleic Acids (UNA)
3. Unlocked Nucleic Acids (UNA)
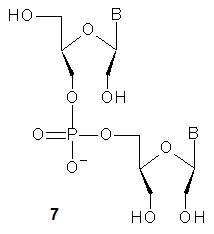
In 1995, Wengel and collaborators described the synthesis of DNA oligonucleotides with 1–3 acyclic unlocked thymidine nucleotides (7, B = Thy, a dinucleotide is shown), that have an OH group at the 2’ position, yet lack a covalent bond between the 2’ and 3’ carbon atoms [6].The unlocked nucleoside was obtained in 88% yield and was converted into the phosphoramidite monomer in good yield. In 2009, the synthesis of other UNA-nucleosides was published and good yields were reported (B = ABz: 73%, CAc: 64%, GiBu: 63%, U: 79%) [7]][7]. The synthetic route to UNA was further improved by the synthesis of 2’-O-TBDMS monomers [8]][8]. These monomers were used in the automated solid phase synthesis of chimeric DNA-UNA and RNA-UNA oligomers. Further studies showed that the UNA units confer them remarkably high stability in serum [7],[9]][7][9].
A single UNA nucleotide present in a DNA strand decreases the Tm value for the corresponding (UNA-DNA)/DNA duplex by 10 °C [6]. The thermal stability of RNA/DNA and RNA/RNA duplexes containing one or two UNA nucleotides, as well as the effects of mismatches, were investigated using a set of 8 RNA 21-mers [7]. It was found that for duplexes formed with DNA or RNA templates, a single UNA unit in a central position of the RNA strands lowered Tm values by 5–8 °C. When the mismatches were positioned directly opposite a UNA monomer, only little discrimination was observed. Additionally, two UNA monomers can be used to decrease or increase mismatch discrimination depending on their position in the oligomers. This phenomenon was used to detect a breast cancer onco micro -RNA (miR-10b) and distinguish it from two other miRNAs (miR-10a and miR10c) that differed from miR10b by one and two nucleotides, respectively [10]][10].
The CD spectra indicate that a UNA nucleotide can be considered structurally as an RNA mimic, since the incorporation of one or a few UNA monomers into the RNA/RNA duplexes does not change the overall structure compared to that of the unmodified duplex [11]. It was documented that UNA nucleotides interact according to the Watson-Crick base-pairing rules, while the stacking interactions of UNA nucleotides are less favorable than those of RNA units.
The UNA nucleotides have also been studied in terms of their effects on the stability of i-motif and G-quadruplex high-ordered structures [12]. A widely known i-motif sequence is a 22-nt fragment of human telomeric DNA with the sequence 5’-d(CCCTAA)3CCCT). This molecule was engineered to form twelve 22-nt oligomers, each containing a single UNA modification located in oligomers in a "walking mode" [13]. The thermodynamic stability of the i-motif structures was measured in buffers of pH 4.8 and 5.6. It was found that the UNA units, depending on their position, either destabilize (positions C1 or C14) or stabilize (U10, A11, A12) the structure. The recorded CD spectra allow to conclude that the UNA-nucleotides do not cause significant changes in the i-motif structure and folding molecularity.
The influence of UNA nucleotides on the structure and stability of G-quadruplexes was investigated using CD spectroscopy. After analyzing a series of oligomers of the general sequence GGGTTTTTGGGA(T,C)GGGTTTTGGG, it was found that UNA nucleotides present in the loops cause global stabilization of the structure, while those in the stem cause destabilization of the quadruplexes [14]. These observations were relevant to the study of the UNA-modified thrombin-binding aptamer 5’-GGTTGGTGTGGTTGG-3’, which forms an intramolecular antiparallel G-quadruplex that interacts with two thrombin molecules and inactivates only one of them. It was found that an aptamer analog carrying UNA-U at position 7 (a loop position) inhibited blood clotting more than the unmodified compound [9].
Pasternak and co-workers also studied an RE31 aptamer, which is a longer version of the thrombin-binding aptamer 5’-GGTTGGTGTGGTGG-3’ mentioned above. Of the 13 analogs synthesized, 6 oligomers contained a single UNA unit (A, C, G and U), one oligomer had 3 UNA nucleotides, other compounds contained one UNA and multiple LNA nucleotides and one oligomer was modified with 14 β-L-RNA units [15]. The greatest increase in thermal stability of RE31 was observed for oligomers labeled with the LNA units present in the duplex fragment or with a combination of LNAs in the helix and UNAs at position 15. Additionally, ternary substitution of T11, T15 and T20 by UNA-U resulted in a 5.4 °C increase in Tm.
Several papers have been published on the use of UNA nucleotides to improve the potency of siRNA oligomers, some siRNAs with UNA units proved to be remarkably stable (e.g., in mice) and, in contrast to unmodified siRNAs, they efficiently induced knock -down of eGFP expression in a pancreas tumor xenograft model [16]. However, the great complexity of the RNAi mechanism confronts researchers with many factors that must be considered to properly identify the effects of UNA nucleotides and other chemical modifications on siRNA activity [17],[18][17][18].
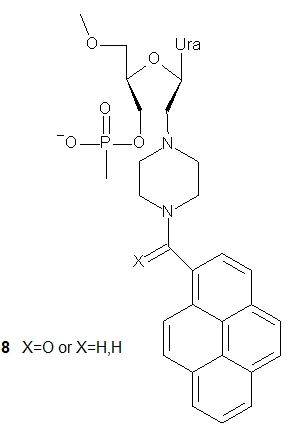
UNA-nucleotides have also been tested as fluorescently labeled compounds to be used as sensitive probes in molecular biology or nanotechnology. Two pyrene-modified UNA monomers were synthesized and incorporated into 21-mer DNA oligonucleotides [19]. The reported units differed in a linker between the 2’-(piperazin-N-yl) moiety and the pyrene molecule, which was either an amide (8, X = O) or an alkylamine (8, X = H,H). It was found that both pyrene-UNA-U units increased the duplex stability compared to UNA monomers, although the corresponding unmodified reference duplexes were still the most stable. Thermodynamic studies showed that they were able to discriminate mismatches well. Pyrene excimer emission was observed in single-stranded oligonucleotides containing three pyrene-UNA-U units, which disappeared after hybridization with DNA. Interestingly, for the first time, the fluorescence intensities of oligomers containing acyclic pyrene-nucleotides were found to increase upon hybridization with DNA or RNA templates. The unit containing the alkylamino linker (8, X = H,H) was used to construct a quencher-free molecular beacon that showed an increase in pyrene excimer emission of more than 900% upon hybridization of a fully complementary RNA target. This probe enabled the detection of RNA in H1299 cells [20].
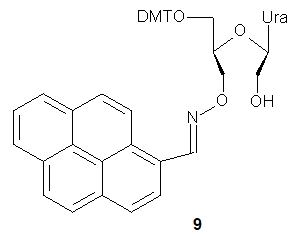
UNA 3’-O-amino-pyrene)-U (9) was used to preparation of oligomers containing this fluorescent pyrene probe. Thermal dissociation studies showed that the presence of 9 in DNA or 2’-OMe RNA oligomers (21-nt long) resulted in increased thermal stability of duplexes with a DNA complement. Fluorescence spectroscopy studies indicated that the pyrenes were located in the minor grooves of the duplexes, and these results were confirmed by molecular modeling, CD, and UV/Vis absorption experiments. Pyrene–pyrene excimer signals were observed for oligomers bearing two or three pyrene-labeled units. Upon hybridization with the complementary DNA strand, the excimer signals disappeared, allowing such constructs to be used for fluorescence-based detection of mismatched hybridization.
- Glycol and Butyl Nucleic Acids (GNA and BuNA)
4. Glycol and Butyl Nucleic Acids (GNA and BuNA)
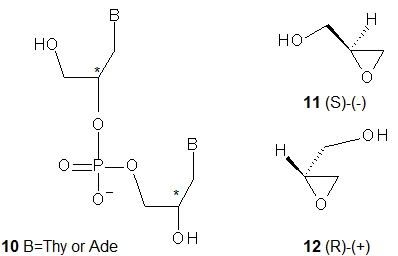
Structurally simple and synthetically relatively easily accessible acyclic glycol nucleic acids (GNA; 10, B = Thy or Ade, a dinucleotide is shown) were obtained by Meggers and coworkers [21]. Note that unlike FNA and UNA, a GNA scaffold contains stereogenic centers (marked with the asterisks in 10) and GNA units can exist in R and S forms. Fortunately, the enantiomerically pure substrates, i.e., (S)-(−)-glycidol (11) and (R)-(+)-glycidol (12), are commercially available. Carefully designed GNAs allowed the study of the formation of anti-parallel and parallel duplexes. It was found that uniformly modified GNA oligomers form highly stable (Tm = 63 °C) anti-parallel helical duplex structures, while the melting curves of the oligomers intended to form a parallel duplex showed no transition. Notably, a single mismatch in the middle of the sequence significantly reduced the stability of the duplex (Tm = 55 °C). It was found that (S)-GNA and its Watson-Crick -paired (R)-GNA counterpart neither hybridize with each other nor form stable duplexes with DNA. However, initial results showed that (S)-GNA 2’-ATTTTAAATATAATAATT-3’ (and not (R)-GNA) associates with RNA (Tm = 35 °C). Analysis of the thermal stability of a GNA/GNA duplex formed by 3’-CACATTATTGTTGTA-2’ with 2’-GTGTAATAACAACAT-3’ revealed a Tm of 71 °C compared to 46 °C for a normal DNA duplex of the same sequence.
To characterize the reasons for the high stability of certain GNA duplexes, the values of ΔG (Gibbs free energy), ΔH (change in enthalpy) and ΔS (change in entropy) for the formation of three duplexes were determined from melting experiments [22]. The calculated thermodynamic parameters indicate that the entropic toll for GNA is much lower than that for DNA, so there must be a strong conformational preorganization of the GNA strands. There were also other results indicating particularly favorable stacking interactions in GNA duplexes. However, whereas in B-DNA the base–base stacking interactions predominantly operate in an intra-strand fashion, in GNA duplexes, these energetically beneficial contacts are between the bases of the opposite strands.
In the field of molecular biology, GNA oligomers have been tested as templates for template-dependent synthesis of DNA using the polymerases Taq, Bst, DeepVent (exo-), Therminator, Sequenase and SuperScript II DNA [23]. For most polymerases, the primer extension process stalled after elongation of the primer by two or three deoxynucleotides. However, the Bst DNA polymerase catalyzed full-length DNA synthesis on a dodecamer GNA template. In another study, the ability of various DNA polymerases (vide supra) to use GNA triphosphates as substrates for GNA synthesis on DNA templates was investigated [24]. It was found that the Therminator DNA polymerase was able to extend the primer by incorporating two GNA units, whereas extension with up to five GNA nucleotides was much less efficient.
In another type of experiment, a GNA-T unit was introduced into a molecule of the thrombin-binding aptamer mentioned above, the important part of which is a G-quadruplex [25]. The GNA-T nucleotide was introduced at position 3, 4, 7, 9, 12 or 13, and in all cases, these substitutions decreased the melting temperature (by approximately 10 °C), except at position 7, where Tm increased by 5 °C. However, no data were shown on the effects of these substitutions on the activity of the aptamer. The effect of GNA substitution on the potency of the siRNA probe was also investigated. Modification of an siRNA with (S)-GNA resulted in higher in vitro efficacy than analogs with (R)-GNA. Notably, duplexes modified with GNA were able to maintain their efficacy in vivo when injected subcutaneously into mice.
Several reports indicate a potential application of GNA oligomers in nanotechnology. Since GNA provides easy access to both left- and right-handed helical geometries, these molecules can potentially be used to construct nanostructures with unique topologies. Chaput and co-workers published a report on the synthesis of two 4-helix junctions (4HJ) with mirror-image symmetry using (R)- or (S)-GNA monomers [26]. Spectra differed significantly from spectra typical of natural DNA and RNA helices, suggesting that the GNA helices have a unique helical structure. Melting experiments showed that the GNA 4HJ was significantly more stable than the corresponding DNA 4HJ (76 vs. 37 °C).
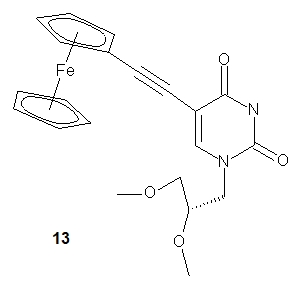
More recently, Kowalski and coworkers published the synthesis of a ferrocenyl-labeled analog of the nucleoside GNA-U (GNA-FcU, 13). The authors suggest a future application of GNA-FcU in the synthesis of redox-active self-assembling “molecular wires”.
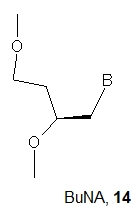
Apart from the acyclic structure, GNA nucleotides differ from DNA and RNA units in the number of atoms present in the backbone units. There are 5 atoms in GNA (O3'-C3'-C2'-O2'-P) and six atoms in natural nucleotides (O5'-C5'-C4'-C3-O3'-P). It has been suggested that the difference in linker length is one reason why GNA oligomers do not hybridize with DNA. Therefore, it was tempting to make a homolog of GNA with six atoms in the backbone, and this was carried out done with the synthesis of Butyl Nucleic Acids (BuNA, 14) [27]. Ten fully modified (eight of them with a DNA nucleoside at the 3’-end) and eight partially modified oligomers were synthesized. It was found that (S)-butyl nucleic acids (BuNA) form duplexes with their complementary strands, but the observed thermal stability was lower than that for corresponding DNA duplexes. Thus, increasing the backbone length by one carbon atom resulted in a decrease in the melting temperature of (S)-BuNA duplexes compared to GNA duplexes. Moreover, the CD spectra showed that (S)-BuNA duplexes adopt the A-like conformation and form a left-handed helical structure. The incorporation of (S)-BuNA nucleotides into DNA strands resulted in destabilized duplexes. The primer extension studies on fully modified (S)-BuNA strands and (S)-BuNA-substituted DNA were carried out using the polymerases Bst, Bsu, Taq and Therminator. No extension was observed with fully modified primers, but when (S)-BuNA -substituted DNA strands were used, full extension of the modified primers was observed.
- Threoninol Nucleic Acids (TNA)
5. Threoninol Nucleic Acids (TNA)
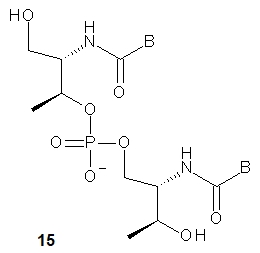
The nucleoside precursors of acyclic threoninol nucleic acid (D-aTNA, 15, a dinucleotide is shown) were prepared using DMT-protected D-threoninol (16, Scheme 2), except for the thymine derivative, for which unprotected D-threoninol 17 was used [28]. The amino groups were used to attach N1-carboxymethyl derivatives of the nucleobases CBz or T, or N9-carboxymethyl derivatives of ABz or GiBu under conditions known from peptide chemistry. The product containing a thymine nucleobase was tritylated and all four DMT-protected nucleosides 18 were routinely phosphitylated to yield the corresponding D-aTNA monomers 19.
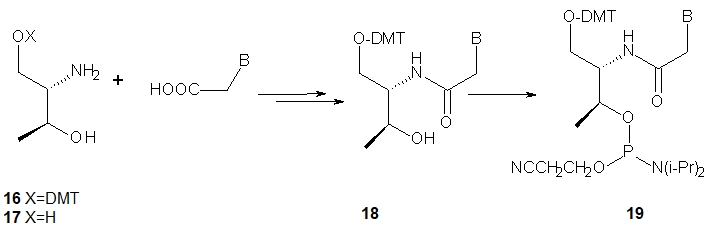
Scheme 2. Synthesis of D-aTNA monomers
19
.
Using the monomers 19, seven oligomers (each 8 -nt long) were synthesized on an automated DNA synthesizer. Melting experiments showed that the D-aTNA oligomers 1’-GCATCAGT-3’ and 3’-CGTAGTCA-1’ formed an unexpectedly stable antiparallel duplex with Tm = 63 °C, compared with analogous DNA and RNA duplexes with 29 and 39 °C, respectively. Notably, a single T:C mismatch significantly lowered the Tm to 49 °C. CD Studies showed that single-stranded D-aTNA oligomers, unlike GNA, do not form a helical preorganized structure. The same group reported the synthesis of L-aTNA using an optimized method analogous to that used in the synthesis of D-aTNA [29]. As expected from the enantiomeric relationship between L- and D-aTNA, the L-aTNAs formed an antiparallel homo-duplex with the same melting temperature (58 °C) as the D-aTNA duplex of the same sequence. However, unlike D-aTNA, L-aTNA not only hybridized quite well with both DNA (Tm = 25–28 °C) and RNA (Tm = 38–41 °C), but the association also occurred in a parallel manner. The crystal structure of an L-aTNA/RNA duplex (3’-GCAGCAGC-1’)/(5’-GCUGCUBrGC-3’) clearly shows right-handed helicity and a Watson-Crick base -pairing [30]. It was shown that L-aTNA is able to block Bsu DNA polymerase by forming a triplex with the DNA template. The triplex formation phenomenon has been used for reversible regulation of polymerase activity and protein expression [31].
Exploration of higher -order structures formed by aTNAs continued and L-aTNA-based G-quadruplex structures were identified [32]. UV-melting, CD, electrophoretic mobility shift assay and fluorescence studies showed that both intermolecular and intramolecular G-quadruplex structures can exist, the latter being more stable. The same group showed that at low pH, cytosine segments in L-aTNA form structures that exhibit the features of the i-motif [33].
L-Threoninol-modified RNA molecules were tested as siRNAs in the RNAi pathway. Two L-aTNA-T units were incorporated at the 3’-termini of the antisense and sense RNA strands of siRNA targeting the Renilla luciferase mRNA [34]. The siRNA containing L-aTNA-(TT) in the 3’-overhang of the antisense strand showed 30% higher gene silencing than the siRNA carrying two natural thymidine residues. A time-course analysis of the siRNAs over five days showed that the siRNA containing two L-threoninol units at both 3'-ends exhibited higher gene silencing activity than the native siRNA from day 3. This duplex showed exceptional stability in human serum, with ~5% of the original siRNA still intact after 8 hours, whereas the native congener was completely degraded after approximately 60 minutes of incubation. Furthermore, the absence of cytotoxicity of the L-threoninol modification was determined by the MTT assay. The siRNA containing L-aTNA-(TT) activates IL-1β production less efficiently than the unmodified siRNA, so the L-threoninol-thymine monomer may potentially be used to attenuate the activation of the immune response.
In the field of analytical biochemistry, DNA probes containing 2–6 D-threoninol-based units with a perylene moiety attached (instead of a nucleobase) have been used in experiments for fluorescence monitoring of DNA [35]. This methodology was extended to the detection of RNA in cell lysates [36].
- Serinol Nucleic Acids (SNA)
6. Serinol Nucleic Acids (SNA)
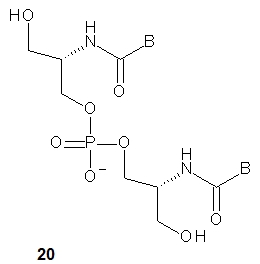
Compared to aTNA, the acyclic flexible scaffold of serinol nucleic acids (20, a dinucleotide is shown) does not contain a methyl group (in the position next to the amino group), which gives rise to the phenomenon of two stereogenic centers in the aTNA unit [37]. Thus, the chirality of the SNA molecule synthesized from four SNA monomers depends only on its sequence. If the sequence is palindromic (e.g., ATTGCGTTA or pATTGCGTTAp), the entire molecule is achiral, although there is a carbon atom with four different substituents in each SNA unit except the central one. When a sequence is non-palindromic (e.g., ATTGCGTAA, pATTGCGTTA or ATTGCGTTAp), the molecule is chiral.
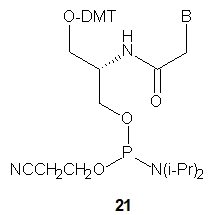
Four chiral SNA-phosphoramidite monomers 21 were synthesized (B=ABz, T, GiBu or CBz) and used for the synthesis of seven SNA-octamers. Their combinations should yield chiral antiparallel, chiral parallel and achiral duplexes. The melting experiments showed that not only were the expected antiparallel SNA homoduplexes formed (Tm = 51 °C), but also the parallel ones, although the latter were much less stable (Tm = 16 °C). Importantly, however, the antiparallel SNA/SNA duplexes were much more stable than the isosequential DNA/DNA (Tm = 23 or 29 °C) or RNA/RNA (Tm = 38 or 39 °C) complexes. Analogously to the previously described L-aTNA/RNA duplex [30] a crystal structure showed that an SNA/RNA duplex adopts right-handed helicity and uses the Watson-Crick base pairing.
The SNA scaffold has been successfully used to prepare molecular beacons (SNA-MB) bearing a fluorophore (perylene or Cy3) and the anthraquinone quencher at opposite ends [38]. The SNA-MBs were resistant to degradation by nuclease and the probes could be used to visualize mRNA in fixed cells.
It was found that SNA-type nucleotides could enhance the efficacy of oligomers acting in the RNA interference process. In the dual luciferase assay, an siRNA duplex and its analogs modified with 1, 3, 5, or 7 SNA nucleotides at each of the sense strands or at the 3' end of the antisense strand were prepared [39].
The aforementioned phenomenon that SNA molecules of palindromic sequences are achiral was the basis for the construction of a nanowire that functions as a helical amplification system [40]. The authors suggest that the SNA nanowire system could be used to detect complementary DNA or RNA fragments, as SNA can recognize natural nucleic acids. Other biomolecules (proteins, sugars) could also be detected if these molecules could induce helicity in the SNA nanostructures.
Funding: This work was financially supported by statutory funds of the Centre of Molecular and Macromolecular Studies, Polish Academy of Sciences, Łódź, Poland.
Conflicts of Interest: The authors declare no conflict of interest.
References
[1]. Leumann, C. J. DNA analogues: from supramolecular principles to biological properties. Bioorg. Med. Chem. 2002, 10, 841–854.
[2]. Joyce, G. F.; Schwartz, A. W.; Miller, S. L.; Orgel L. E. The case for an ancestral genetic system involving simple analogues of the nucleotides. Proc. Natl. Acad. Sci. U.S.A. 1987, 84, 4398–440.
[3] Schneider, K.C.; Benner, S.A. Oligonucleotides containing flexible nucleoside analogs. J. Am. Chem. Soc. 1990, 112, 453–455.
[4]. Merle, Y.; Bonneil, E.; Merle, L.; Saggi, J.; Szemzo, A. Acyclic oligonucleotide analogues. Int. J. Biol. Macromol. 1995, 17, 239–246.
[5]. Chaput, J. C.; Switzer C. Nonenzymatic oligomerization on templates containing phosphodiester-linked acyclic glycerol nucleic acid analogues. J. Mol. Evol . 2000, 51, 464–470.
[6]. Nielsen, P.; Dreiøe, L. H.; Wengel, J. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides: introduction of three novel analogues and a summary. Bioorg. Med. Chem. 1995, 3, 19–28.
[7]. Langkjær, N.; Pasternak, A.; Wengel, J. UNA (unlocked nucleic acid): a flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg. Med. Chem. 2009, 17, 5420–5425.
[8]. Mangos, M. M.; Min, K.-L.; Viazovkina, E.; Galarneau, A.; Elzagheid, M. J.; Parniak, M. A.; Damha, M. J. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2'F-ANA constructs containing acyclic nucleotide inserts. J. Am. Chem. Soc. 2003, 125, 654-661.
[9]. Jensen, T. B.; Langkjær, N.; Wengel, J. Unlocked nucleic acid (UNA) and UNA derivatives: thermal denaturation studies. Nucleic Acids Symp. Ser. (Oxf), 2008, 52, 133-134.
[10]. Robertson, N. M.; Toscano, A. E.; LaMantia, V. E.; Salih Hizir, M.; Rana, M.; Balcioglu, M.; Sheng, J.; Yigit, M. V. Unlocked Nucleic Acids for miRNA detection using two dimensional nano-graphene oxide. Biosens. Bioelectron., 2017, 89, 551–557.
[11]. Pasternak, A.; Wengel, J. Thermodynamics of RNA duplexes modified with unlocked nucleic acid nucleotides. Nucleic Acids Res . 2010, 38, 6697-6706.
[12]. Guéron, M.; Leroy, J.-L. The i-motif in nucleic acids. Curr. Opin. Struct. Biol . 2000, 10, 326-331.
[13]. Pasternak, A.; Wengel, J. Modulation of i-motif thermodynamic stability by the introduction of UNA (unlocked nucleic acid) monomers. Bioorg. Med. Chem. Lett . 2011, 21, 752-755.
[14]. Agarwal, T.; Kumar, S.; Maiti, S. Unlocking G-quadruplex: Effect of unlocked nucleic acid on G-quadruplex stability. Biochimie 2011, 93, 1694-1700.
[15]. Kotkowiak, W.; Wengel, J.; Scotton, C. J.; Pasternak, A. Improved RE31 Analogues Containing Modified Nucleic Acid Monomers: Thermodynamic, Structural, and Biological Effects. J. Med. Chem. 2019, 62, 2499-2507.
[16]. Laursen, M. B.; Pakula, M. M.; Gao, S.; Fluiter, K.; Mook, O. R.; Baas, F.; Langklær, N.; Wengel, S. L.; Wengel, J.; Kjemsab, J.; Bramsen, J. B. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol. BioSyst. 2010, 6, 862-870.
[17]. Kenski, D. M.; Cooper, A. J.; Li, J. J.; Willingham, A. T.; Haringsma, H. J.; Young, T. A.; Kuklin, N. A.; Jones, J. J.; Cancilla, M. T.; McMasters, D. R.; Mathur, M.; Sachs, A. B.; Flanagan. W. M. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5'-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res. 2010, 38, 660-671.
[18]. Vaish, N.; Chen, F.; Seth, S.; Fosnaugh, K.; Liu, Y.; Adami, R.; Brown, T.; Chen, Y.; Harvie, P.; Johns, R.; Severson, G.; Granger, B.; Charmley, P.; Houston, M.; Templin, M. V.; Polisky, P. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011, 39, 1823-1832.
[19]. Karlsen, K. K.; Pasternak, A.; Jensen, T. B.; Wengel, J. Pyrene-modified unlocked nucleic acids: synthesis, thermodynamic studies, and fluorescent properties. ChemBioChem 2012, 13, 590-601.
[20]. Karlsen, K. K.; Okholm, A.; Kjems, J.; Wengel, J. A quencher-free molecular beacon design based on pyrene excimer fluorescence using pyrene-labeled UNA (unlocked nucleic acid). Bioorg. Med. Chem. 2013, 21, 6186-6190.
[21]. Zhang, L.; Peritz, A.; Meggers, E. A simple glycol nucleic acid. J. Am. Chem. Soc. 2005, 127, 4174-4175.
[22]. Schlegel, M. K.; Xie, X.; Zhang, L.; Meggers. E. Insight into the high duplex stability of the simplified nucleic acid GNA. Angew. Chem. Int. Ed. 2009, 48, 960-963.
[23]. Tsai, Ch-H.; Chen, J.; Szostak, J. W. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 14598-14603.
[24]. Chen, J. J.; Tsai, Ch.-H.; Cai, X.; Horhota, A. T.; McLaughlin, L. W.; Szostak, J. W. Enzymatic primer-extension with glycerol-nucleoside triphosphates on DNA templates. PLoS One 2009, 4, e4949.
[25]. Aviñó, A.; Mazzini, S.; Fàbrega, C.; Peñalver, P.; Gargallo, R.; Morales, J. C.; Eritja, R. The effect of l-thymidine, acyclic thymine and 8-bromoguanine on the stability of model G-quadruplex structures. Biochim. Biophys. Acta 2017, 1861, 1205-1212.
[26]. Zhang, R. S.; McCullum, E. O.; Chaput, J. C. Synthesis of Two Mirror Image 4-Helix Junctions Derived from Glycerol Nucleic Acid. J. Am. Chem. Soc. 2008, 130, 5846-5847.
[27]. Kumar, V.; Gore, K. R.; Pradeepkumar, P. I.; Kesavan, V. Design, synthesis, biophysical and primer extension studies of novel acyclic butyl nucleic acid (BuNA). Org. Biomol. Chem. 2013, 11, 5853-5865.
[28]. Asanuma, H.; Toda, T.; Murayama, K.; Liang, X.; Kashida, H. Unexpectedly Stable Artificial Duplex from Flexible Acyclic Threoninol. J. Am. Chem. Soc. 2010, 132, 14702-14703.
[29]. Murayama, K.; Kashida, H.; Asanuma, H. Acyclic L-threoninol nucleic acid (L-aTNA) with suitable structural rigidity cross-pairs with DNA and RNA. Chem. Commun. 2015, 51, 6500-6503.
[30]. Kamiya, Y.; Slatoh, T.; Kodama, A.; Suzuki, T.; Murayama, K.; Kashida, H.; Uchiyama, S.; Kato, K.; Asanuma, H. Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA. Commun. Chem. 2020, 3, 156.
[31]. Nguyen, T. J. D.; Manuguerra, I.; Kumar, V.; Gothelf, K. V. Toehold-Mediated Strand Displacement in a Triplex Forming Nucleic Acid Clamp for Reversible Regulation of Polymerase Activity and Protein Expression. Chem. Eur. J . 2019, 25, 12303–12307.
[32]. Kumar, V.; Gothel, K. V. Synthesis and biophysical properties of (L)-aTNA based G-quadruplexes. Org. Biomol. Chem. 2016, 14, 1540-1544.
[33]. Kumar, V.; Nguyen, T. J. D.; Palmfeldt, J.; Gothelf, K. V. Formation of i-motifs from acyclic (L)-threoninol nucleic acids. Org. Biomol. Chem. 2019, 17, 7655–7659.
[34] Alagia, A.; Terrazas, M.; Eritja, R. RNA/aTNA Chimeras: RNAi Effects and Nucleases Resistance of Single and Double Stranded RNAs. Molecules 2014, 19, 17872-17896.
[35]. Asanuma, H.; Akahane, M.; Kondo, N.; Osawa, T.; Kato, T.; Kashida, H. Quencher-free linear probe with multiple fluorophores on an acyclic scaffold. Chem. Sci. 2012, 3, 3165–3169.
[36]. Asanuma, H.; Akahane, M.; Niwa, R.; Kashida, H.; Kamiya, Y. Highly Sensitive and Robust Linear Probe for Detection of mRNA in Cells. Angew. Chem. Int. Ed. 2015, 54, 4315–4319.
[37]. Kashida, H.; Murayama, K.; Toda, T.; Asanuma, H. Control of the Chirality and Helicity of Oligomers of Serinol Nucleic Acid (SNA) by Sequence Design. Angew. Chem. Int. Ed. 2011, 50, 1285 –1288.
[38]. Murayama, K.; Kamiya, Y.; Kashida, H.; Asanuma, H. Ultrasensitive Molecular Beacon Designed with Totally Serinol Nucleic Acid (SNA) for Monitoring mRNA in Cells. ChemBioChem 2015, 16, 1298 –1301.
[39]. Kamiy, Y.; Takai, J.; Ito, H.; Murayama, K.; Kashida, H.; Asanuma, H. Enhancement of Stability and Activity of siRNA by Terminal Substitution with Serinol Nucleic Acid (SNA). ChemBioChem 2014, 15, 2549 – 2555.
[40]. Kashida, H.; Nishikawa, K.; Shi, W.; Miyagawa, T.; Yamashita, H.; Abe, M.; Asanuma, H. A helical amplification system composed of artificial nucleic acids. Chem. Sci. 2021, 12, 1656–1660.
References
- Leumann, C. J. DNA analogues: from supramolecular principles to biological properties. Bioorg. Med. Chem. 2002, 10, 841–854.
- Joyce, G. F.; Schwartz, A. W.; Miller, S. L.; Orgel L. E. The case for an ancestral genetic system involving simple analogues of the nucleotides. Proc. Natl. Acad. Sci. U.S.A. 1987, 84, 4398–440.
- Schneider, K.C.; Benner, S.A. Oligonucleotides containing flexible nucleoside analogs. J. Am. Chem. Soc. 1990, 112, 453–455.
- Merle, Y.; Bonneil, E.; Merle, L.; Saggi, J.; Szemzo, A. Acyclic oligonucleotide analogues. Int. J. Biol. Macromol. 1995, 17, 239–246.
- Chaput, J. C.; Switzer C. Nonenzymatic oligomerization on templates containing phosphodiester-linked acyclic glycerol nucleic acid analogues. J. Mol. Evol . 2000, 51, 464–470.
- Nielsen, P.; Dreiøe, L. H.; Wengel, J. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides: introduction of three novel analogues and a summary. Bioorg. Med. Chem. 1995, 3, 19–28.
- Langkjær, N.; Pasternak, A.; Wengel, J. UNA (unlocked nucleic acid): a flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg. Med. Chem. 2009, 17, 5420–5425.
- Mangos, M. M.; Min, K.-L.; Viazovkina, E.; Galarneau, A.; Elzagheid, M. J.; Parniak, M. A.; Damha, M. J. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2'F-ANA constructs containing acyclic nucleotide inserts. J. Am. Chem. Soc. 2003, 125, 654-661.
- Jensen, T. B.; Langkjær, N.; Wengel, J. Unlocked nucleic acid (UNA) and UNA derivatives: thermal denaturation studies. Nucleic Acids Symp. Ser. (Oxf), 2008, 52, 133-134.
- Robertson, N. M.; Toscano, A. E.; LaMantia, V. E.; Salih Hizir, M.; Rana, M.; Balcioglu, M.; Sheng, J.; Yigit, M. V. Unlocked Nucleic Acids for miRNA detection using two dimensional nano-graphene oxide. Biosens. Bioelectron., 2017, 89, 551–557.
- Pasternak, A.; Wengel, J. Thermodynamics of RNA duplexes modified with unlocked nucleic acid nucleotides. Nucleic Acids Res . 2010, 38, 6697-6706.
- Guéron, M.; Leroy, J.-L. The i-motif in nucleic acids. Curr. Opin. Struct. Biol . 2000, 10, 326-331.
- Pasternak, A.; Wengel, J. Modulation of i-motif thermodynamic stability by the introduction of UNA (unlocked nucleic acid) monomers. Bioorg. Med. Chem. Lett . 2011, 21, 752-755.
- Agarwal, T.; Kumar, S.; Maiti, S. Unlocking G-quadruplex: Effect of unlocked nucleic acid on G-quadruplex stability. Biochimie 2011, 93, 1694-1700.
- Kotkowiak, W.; Wengel, J.; Scotton, C. J.; Pasternak, A. Improved RE31 Analogues Containing Modified Nucleic Acid Monomers: Thermodynamic, Structural, and Biological Effects. J. Med. Chem. 2019, 62, 2499-2507.
- Laursen, M. B.; Pakula, M. M.; Gao, S.; Fluiter, K.; Mook, O. R.; Baas, F.; Langklær, N.; Wengel, S. L.; Wengel, J.; Kjemsab, J.; Bramsen, J. B. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol. BioSyst. 2010, 6, 862-870.
- Kenski, D. M.; Cooper, A. J.; Li, J. J.; Willingham, A. T.; Haringsma, H. J.; Young, T. A.; Kuklin, N. A.; Jones, J. J.; Cancilla, M. T.; McMasters, D. R.; Mathur, M.; Sachs, A. B.; Flanagan. W. M. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5'-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res. 2010, 38, 660-671.
- Vaish, N.; Chen, F.; Seth, S.; Fosnaugh, K.; Liu, Y.; Adami, R.; Brown, T.; Chen, Y.; Harvie, P.; Johns, R.; Severson, G.; Granger, B.; Charmley, P.; Houston, M.; Templin, M. V.; Polisky, P. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011, 39, 1823-1832.
- Karlsen, K. K.; Pasternak, A.; Jensen, T. B.; Wengel, J. Pyrene-modified unlocked nucleic acids: synthesis, thermodynamic studies, and fluorescent properties. ChemBioChem 2012, 13, 590-601.
- Karlsen, K. K.; Okholm, A.; Kjems, J.; Wengel, J. A quencher-free molecular beacon design based on pyrene excimer fluorescence using pyrene-labeled UNA (unlocked nucleic acid). Bioorg. Med. Chem. 2013, 21, 6186-6190.
- Zhang, L.; Peritz, A.; Meggers, E. A simple glycol nucleic acid. J. Am. Chem. Soc. 2005, 127, 4174-4175.
- Schlegel, M. K.; Xie, X.; Zhang, L.; Meggers. E. Insight into the high duplex stability of the simplified nucleic acid GNA. Angew. Chem. Int. Ed. 2009, 48, 960-963.
- Tsai, Ch-H.; Chen, J.; Szostak, J. W. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 14598-14603.
- Chen, J. J.; Tsai, Ch.-H.; Cai, X.; Horhota, A. T.; McLaughlin, L. W.; Szostak, J. W. Enzymatic primer-extension with glycerol-nucleoside triphosphates on DNA templates. PLoS One 2009, 4, e4949.
- Aviñó, A.; Mazzini, S.; Fàbrega, C.; Peñalver, P.; Gargallo, R.; Morales, J. C.; Eritja, R. The effect of l-thymidine, acyclic thymine and 8-bromoguanine on the stability of model G-quadruplex structures. Biochim. Biophys. Acta 2017, 1861, 1205-1212.
- Zhang, R. S.; McCullum, E. O.; Chaput, J. C. Synthesis of Two Mirror Image 4-Helix Junctions Derived from Glycerol Nucleic Acid. J. Am. Chem. Soc. 2008, 130, 5846-5847.
- Kumar, V.; Gore, K. R.; Pradeepkumar, P. I.; Kesavan, V. Design, synthesis, biophysical and primer extension studies of novel acyclic butyl nucleic acid (BuNA). Org. Biomol. Chem. 2013, 11, 5853-5865.
- Asanuma, H.; Toda, T.; Murayama, K.; Liang, X.; Kashida, H. Unexpectedly Stable Artificial Duplex from Flexible Acyclic Threoninol. J. Am. Chem. Soc. 2010, 132, 14702-14703.
- Murayama, K.; Kashida, H.; Asanuma, H. Acyclic L-threoninol nucleic acid (L-aTNA) with suitable structural rigidity cross-pairs with DNA and RNA. Chem. Commun. 2015, 51, 6500-6503.
- Kamiya, Y.; Slatoh, T.; Kodama, A.; Suzuki, T.; Murayama, K.; Kashida, H.; Uchiyama, S.; Kato, K.; Asanuma, H. Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA. Commun. Chem. 2020, 3, 156.
- Nguyen, T. J. D.; Manuguerra, I.; Kumar, V.; Gothelf, K. V. Toehold-Mediated Strand Displacement in a Triplex Forming Nucleic Acid Clamp for Reversible Regulation of Polymerase Activity and Protein Expression. Chem. Eur. J . 2019, 25, 12303–12307.
- Kumar, V.; Gothel, K. V. Synthesis and biophysical properties of (L)-aTNA based G-quadruplexes. Org. Biomol. Chem. 2016, 14, 1540-1544.
- Kumar, V.; Nguyen, T. J. D.; Palmfeldt, J.; Gothelf, K. V. Formation of i-motifs from acyclic (L)-threoninol nucleic acids. Org. Biomol. Chem. 2019, 17, 7655–7659.
- Alagia, A.; Terrazas, M.; Eritja, R. RNA/aTNA Chimeras: RNAi Effects and Nucleases Resistance of Single and Double Stranded RNAs. Molecules 2014, 19, 17872-17896.
- Asanuma, H.; Akahane, M.; Kondo, N.; Osawa, T.; Kato, T.; Kashida, H. Quencher-free linear probe with multiple fluorophores on an acyclic scaffold. Chem. Sci. 2012, 3, 3165–3169.
- Asanuma, H.; Akahane, M.; Niwa, R.; Kashida, H.; Kamiya, Y. Highly Sensitive and Robust Linear Probe for Detection of mRNA in Cells. Angew. Chem. Int. Ed. 2015, 54, 4315–4319.
- Kashida, H.; Murayama, K.; Toda, T.; Asanuma, H. Control of the Chirality and Helicity of Oligomers of Serinol Nucleic Acid (SNA) by Sequence Design. Angew. Chem. Int. Ed. 2011, 50, 1285 –1288.
- Murayama, K.; Kamiya, Y.; Kashida, H.; Asanuma, H. Ultrasensitive Molecular Beacon Designed with Totally Serinol Nucleic Acid (SNA) for Monitoring mRNA in Cells. ChemBioChem 2015, 16, 1298 –1301.
- Kamiy, Y.; Takai, J.; Ito, H.; Murayama, K.; Kashida, H.; Asanuma, H. Enhancement of Stability and Activity of siRNA by Terminal Substitution with Serinol Nucleic Acid (SNA). ChemBioChem 2014, 15, 2549 – 2555.
- Kashida, H.; Nishikawa, K.; Shi, W.; Miyagawa, T.; Yamashita, H.; Abe, M.; Asanuma, H. A helical amplification system composed of artificial nucleic acids. Chem. Sci. 2021, 12, 1656–1660.
