You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Mallika Vijayanathan.
Polycomb repressive complex 2 (PRC2) represents a group of evolutionarily conserved multi-subunit complexes that repress gene transcription by introducing trimethylation of lysine 27 on histone 3 (H3K27me3). PRC2 activity is of key importance for cell identity specification and developmental phase transitions in animals and plants.
- polycomb
- PRC2
- H3K27me3
- evolution
- green lineage
- plant
1. Introduction
DNA in the eukaryotic nucleus winds around octamers of histones, forming nucleosomes, the basic units of chromatin. DNA and histones are subject to chemical modifications, such as methylation, phosphorylation, acetylation, and others, which are instructive for or correlate with chromatin structure. Remodeling of chromatin structure between more open (accessible) or more compact (inaccessible) states by chromatin-modifying and -remodeling complexes governs the distribution of DNA in the nuclear space and allows for gene activation or repression, respectively.
Among crucial modulators of chromatin structure are polycomb group (PcG) proteins, which form multi-subunit polycomb repressive complexes (PRCs) (reviewed in the work of [1,2,3,4,5][1][2][3][4][5]). Major PRCs are histone-modifying complexes that confer different and even counteracting enzymatic activities, which mediate gene repression. In animals and plants, PRC1 is an E3 ubiquitin ligase complex that catalyzes histone 2A lysine ubiquitination (H2AKub: K118 in Drosophila, K119 in vertebrates, and K121 in plants) [6[6][7][8],7,8], and PRC2 is a histone methyltransferase (HMT) complex that catalyzes histone 3 lysine 27 methylation (H3K27me) [9]. Some PRCs catalyze H2Aub removal. In animals, the PR-DUB (polycomb repressive deubiquitinase) complex catalyzes histone H2A deubiquitination, which contributes to gene repression (reviewed in the work of [10,11][10][11]). Similarly, in plants, H2A deubiquitinases UBP12 and UBP13 are implicated in polycomb repression [12[12][13],13], but the composition of associated protein complexes remains enigmatic. Additional PRCs have been described in animals, such as Drosophila pleiohomeotic repressive complex (PhoRC) that does not confer enzymatic activity ([14], reviewed in the work of [15]). As histone modifications introduced by PRCs are heritable during mitotic cell divisions (reviewed in the work of [16,17,18][16][17][18]), PRCs provide an epigenetic memory system required for stable cell identity, for an adequate response to external cues, and even for stable repression of genomic repeats (reviewed in the work of [19]). In line with its function in cell identity maintenance in animals, PRC2 dysfunction is frequently associated with cancer development and PRC2 is a potent target for anticancer therapy (reviewed in the work of [20,21,22][20][21][22]).
PRC1 and PRC2 are conserved in animal and plant models. PRC1 was long considered to be animal specific since a homolog of Polycomb (Pc), the Drosophila PRC1-defining protein subunit [23], is missing in plants. Later, orthologs of PRC1 catalytic subunits RING1 (Really Interesting New Gene 1) and BMI1 (B cell-specific Moloney murine leukemia virus integration site 1) were identified in plants and worms [24]. RING1 and BMI1 were shown to be indispensable for plant development and required for H2Aub [7,25,26][7][25][26]. RING1 and BMI1 orthologs, as well as plant-specific PRC1 subunits, have been found in different plants species ([27], reviewed in the work of [28]). Hence, PRC1 is present in plants, although its core composition differs from animals and may have originated through convergent evolution (reviewed in the work of [28,29,30][28][29][30]). In contrast to PRC1, PRC2 core composition and H3K27me activity are well conserved throughout eukaryotic lineages, and PRC2 is hypothesized to have emerged through divergent evolution [31]. The evolution of PRC2 has been shaped by genome duplication and subfunctionalization, and the number of different PRC2 complexes tends to rise with increasing body plan complexity (Figure 1) ([32], reviewed in the work of [15,33,34][15][33][34]). In animals, PRC2 catalyzes H3K27 mono, di, and trimethylation (i.e., H3K27me1, H3K27me2, H3K27me3), but in flowering plants, it catalyzes H3K27me3 ([35,36][35][36], reviewed in the work of [9,18][9][18]). In model species of both animals and plants, H3K27me3 is largely associated with transcriptional silencing of developmental genes ([37[37][38],38], reviewed in the work of [39,40][39][40]). PRC2 composition, its biochemical and developmental functions are well studied in animal and in flowering plant model species, and weauthors refer to recent reviews for detailed information [41,42,43,44,45,46,47,48,49][41][42][43][44][45][46][47][48][49].
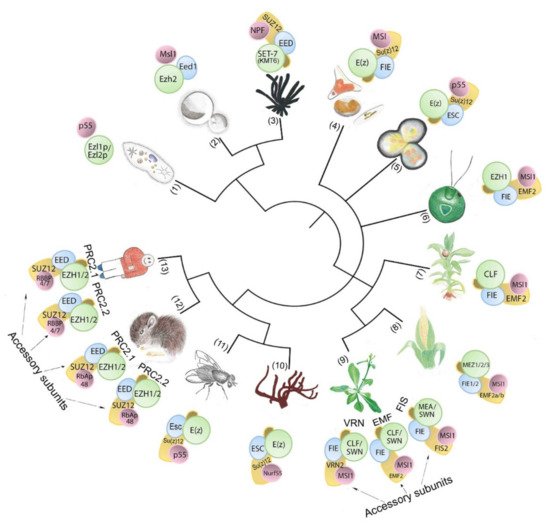
Figure 1. PRC2-core subunit diversity in unicellular and multicellular organisms. The number of subunit homologs and putative PRC2 complexity hypothetically increases with body plan complexity. Numbers in brackets indicate species (1) Paramecium tetraurelia; (2) Cryptococcus neoformans; (3) Neurospora crassa; (4) Phaeodactylum tricornutum; (5) Cyanidioschyzon merolae; (6) Chlamydomonas reinhardtii; (7) Physcomitrium patens; (8) Zea mays; (9) Arabidopsis thaliana; (10) Amphimedon queenslandica; (11) Drosophila melanogaster; (12) Mus musculus; (13) Homo sapiens..
2. Features of PRC2 Core Composition and Function Are Conserved in Animal and Plant Models
The core of PRC2 is generally composed of four protein subunits that are conserved in multicellular eukaryotic model organisms (Figure 1, Table 1). The four subunits enhancer of zeste (E(z)), suppressor of zeste 12 (Su(z)12), extra sex combs (Esc), and nucleosome remodeling factor (Nurf55, also called p55) are the essential PRC2 components present in D. melanogaster (Figure 2) ([50], reviewed in the work of [15]). E(z) contains the catalytic SET domain (Suppressor of variegation 3–9 (Su(var)3–9), Enhancer of zeste (E(z)), Trithorax (Trx) domain), which is responsible for the HMT activity ([50], reviewed in the work of [51]). Su(z)12 is a VEFS (VRN2-EMF2-FIS2-Su(z)12) domain-containing protein, and Esc and p55/Nurf55 are two WD40 repeat (WDR) domain proteins. Su(z)12, Esc, and Nurf55 are non-catalytic subunits that are crucial for PRC2 catalytic activity ([52,53,54,55][52][53][54][55], reviewed in the work of [56]). PRC2 containing all four core components is 1000 times more active than the E(z) subunit alone [50,57][50][57]. Esc and Su(z)12 play critical roles in stimulating the HMTase activity of E(z) [58[58][59],59], and the loss of Esc impairs global H3K27me in Drosophila embryos [52,57][52][57]. Homologs of Drosophila PRC2 subunits are conserved in mammals, fungi, plants, red algae, and diatoms (Figure 1, Table 1) ([60[60][61][62][63][64][65],61,62,63,64,65], reviewed in the work of [66,67,68,69][66][67][68][69]). In mammals, the homologs of E(z), Su(z)12, Esc, and p55 are EZH1 and EZH2, SUZ12, EMBRYONIC ECTODERM DEVELOPMENT (EED), and RETINOBLASTOMA BINDING PROTEINS (RBBP4 or RBBP7), respectively. Minimal PRC2 in mammals comprises the EZH1/2, EED, and SUZ12 subunits in a 1:1:1 stoichiometry [70], and the trimeric core is responsible for H3K27me1/2/3 (reviewed in the work of [71]). RBBP4 or RBBP7, which display 92% sequence identity, are frequently found together in the same complex, but they may differ functionally, as only RBBP4 is involved in maintaining stem cell identify ([72], reviewed in the work of [73]). Core PRC2 possesses lower activity and/or affinity to target sites unless associated with accessory proteins (reviewed in the work of [56]) that define subtypes of PRC2s ([74], reviewed in the work of [75,76][75][76]). In humans, two PRC2 subtypes are present based on association with accessory proteins, namely PRC2.1 (contains polycomb-like 1–3 (PCL1/2/3), elongin BC and PRC2 (EPOP), PRC2-associated LCOR isoform 1/2 (PALI1/2)), and PRC2.2 (contains jumonji and AT-rich interaction domain-containing 2 (JARID2), adipocyte enhancer-binding protein (AEBP2)) (reviewed in the work of [10]). The molecular structures have been determined for Drosophila, human, mouse, and the fungus Chaetomium thermophilum PRC2 (Figure 2) ([60,77,78,79,80][60][77][78][79][80], reviewed in the work of [34,81,82,83,84][34][81][82][83][84]).
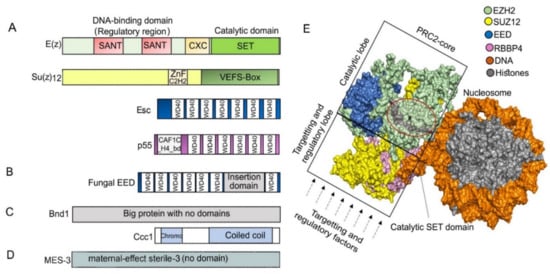
Figure 2. (A) Typical domain architecture of PRC2 core subunits in D. melanogaster. The catalytic subunit E(z) contains SANT (SWI3-ADA2-N-CoR-TFIIIB), CXC (pre-SET domain with C-X(6)-C-X(3)-C-X-C motif), and SET (Su(var)3–9, enhancer of zeste, and trithorax domain) domains, and it catalyzes H3K27me through the SET domain. Su(z)12 contains ZnF_C2H2 (cysteine 2-histidine 2 zinc finger domain) and VEFS (VRN2-EMF2-FIS2-Su(z)12) domain and is responsible for assembling the PRC2 complex, while Esc (WD40 repeat protein) helps to stabilize and enhance E(z) activity. P55 (WD40 repeat protein) can bind to histones and Su(z)12 and is necessary for nucleosome interaction. (B) Most of the studied fungal Esc homologs EED carry a long insertion domain in their C-terminal part. (C) While Su(z)12 is missing, subunits Bnd1 (Big protein with no domains) and Ccc1 (Chromodomain and a coiled coil region) are present in Cryptococcus neoformans. (D) Su(z)12 is missing in C. elegans, but the protein MES-3 (no detectable domains) is a part of the core PRC2. (E) PRC2 core structure. Surface depiction of human PRC2 core complex was created using PyMOL 2.4.1 [85] based on PDB ID: 6WKR [86]; only core subunits are shown. In a catalytically active complex, the EED subunit is encircled by EZH2, and the C-terminal VEFS domain of SUZ12 is sandwiched between the EED and EZH2-SET domain [87]. The region in the circle highlights the lysine binding channel (gray) through which a methyl group is transferred from the cofactor S-adenosylmethionine (SAM)) to the substrate H3 peptides. The catalytic or targeting and regulatory functions of mammalian PRC2 are separated into two regions within the structure: into the top catalytic lobe (containing EZH, VEFS domain of SUZ12 and EED) and the bottom targeting and regulatory lobe (containing RBBP4/7 and N-terminal SUZ12) (reviewed in the works of [42,46,84,88][42][46][84][88]).
Table 1. The distribution of PRC2 core components in different species. (Black ✓ indicates prediction and green ✓ indicates experimental evidence; numbers in brackets indicate the number of homologs identified, ‘?’ indicates that the presence or absence is still unclear, and ‘-’ indicates absence.)
| Supergroups | Eukaryotic Group/Kingdom | Phylum or Class | Species | E(z) | Su(z)12 | Esc | p55 | References | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Archaeplastida | Eukaryota | Rhodophyta | Cyanidioschyzon merolae | ✓ | ✓ | ✓ | ✓ (2) | [63] | ||
| Viridiplantae | Chlorophyta | Ostreococcus lucimarinus | ✓ | ✓ | ✓ | ✓(2) | [89] | |||
| Chlamydomonas reinhardtii | ✓ | ✓ | ✓(2) | ✓(2) | [63,89] | [63][89] | ||||
| Volvox carteri | ✓(2) | ? | ✓ | ✓(2) | [89] | |||||
| Viridiplantae—Embryophyta | Bryophyta-Bryopsida | Physcomitrium patens | ✓ | ✓(3) | ✓ | ✓(2) | [61,62,89,90] | [61][62][89][90] | ||
| Lycophyte-Lycopodiopsida | Selaginella moellendorffii | ✓(2) | ✓ | ✓ | ✓(3) | [89,90,91] | [89][90][91] | |||
| Gymnosperm | Picea abies | ✓ | ? | ? | ? | [92] | ||||
| Angiosperms-Monocot | Oryza sativa | ✓(2) | ✓(2) | ✓(2) | ✓(2) | [91,93,94,95,96] | [91][93][94][95][96] | |||
| Triticum aestivum | ✓(9) | ✓(8) | ✓ (7) | ✓(6) | [97] | |||||
| Zea mays | ✓(3) | ✓(2) | ✓(2) | ✓(5) | [94] | |||||
| Brachypodium distachyon | ✓(2) | ✓(2) | ✓(3) | ✓(4) | [89,98,99] | [89][98][99] | ||||
| Hordeum vulgare | ✓(3) | ✓(3) | ✓ | ✓(2) | [94,100,101] | [94][100][101] | ||||
| Sorghum bicolor | ✓(2) | ✓(3) | ✓(2) | ✓(2) | [94] | |||||
| Angiosperms-Eudicot | Arabidopsis thaliana | ✓(3) | ✓(3) | ✓ | ✓(5) | [102] | ||||
| Chromalveolata | SAR—Stramenopiles | Bacillariophyceae (diatoms) | Phaeodactylum tricornutum | ✓ | ✓ | ✓ | ✓ | [89,103] | [89][103] | |
| Ochrophyta-Phaeophyceae | Ectocarpus | - | - | - | - | [104] | ||||
| SAR—Alveolata | Cilliophora/cilliates | Paramecium tetraurelia | ✓(2) | - | - | ✓ | [31,105,106] | [31][105][106] | ||
| Tetrahymena thermophila | ✓ | ✓ | ✓ | ✓ | [107,108] | [107][108] | ||||
| Opisthokonta | Fungi | Basidiomycota | Cryptococcus neoformans | ✓ | - | ✓ | ✓ | [64,109] | [64][109] | |
| Ascomycota | Fusarium graminearum | ✓ | ✓ | ✓ | ✓ | [109,110] | [109][110] | |||
| Chaetomium thermophilum | ✓ | ✓ | ✓ | ✓ | [79] | |||||
| Neurospora crassa | ✓ | ✓ | ✓ | ✓ | [109, | [ | 111, | 109 | 112] | ][111][112] |
| Saccharomyces cerevisiae | - | - | - | - | [31,75] | [31][75] | ||||
| Filasterea—single-celled eukaryote | Capsaspora | Capsaspora owczarzaki | - | - | - | - | [113] | |||
| Animalia/animals | Porifera | Amphimedon queenslandica | ✓(4) | ✓ | ✓(2) | ✓ | [114] | |||
| Cnidaria/Hydrozoa | Hydra vulgaris | ✓ | ✓ | ✓ | ✓ | [115,116,117, | [116][117 | 118] | [115]][118] | |
| Insecta/insects | Drosophila melanogaster | ✓ | ✓ | ✓ | ✓ | [119,120,121] | [119][120][121] | |||
| Nematoda/nematodes | Caenorhabditis elegans | ✓ | - | ✓ | - | [122, | [122 | 123] | ][123] | |
| Reptilia/reptiles | Anolis carolinensis | ✓ | ✓ | ✓ | ✓ | [124,125,126,127] | [124][125][126][127] | |||
| Mammalia/mammals | Homo sapiens | ✓(2) | ✓ | ✓ | ✓ | [56,60] | [56][60] | |||
| Mus musculus | ✓(2) | ✓ | ✓ | ✓ | [128,129] | [128][129] |
The flowering dicot model plant Arabidopsis thaliana has three E(z) paralogs (CURLY LEAF (CLF), SWINGER (SWN) and MEDEA (MEA)), three Su(z)12 paralogs (EMBRYONIC FLOWER 2 (EMF2), VERNALIZATION 2 (VRN2), and FERTILIZATION INDEPENDENT SEED 2 (FIS2)), one ESC homolog (FERTILIZATION INDEPENDENT ENDOSPERM (FIE)), and five p55 homologs (MULTICOPY SUPPRESSOR OF IRA (MSI1–MSI5)), of which only MSI1 is known to be present in PRC2 ([102[102][130][131],130,131], reviewed in the works of [29,132][29][132]). SWN and CLF play significant roles during vegetative development and phase transitions ([133,134,135,136[133][134][135][136][137][138],137,138], reviewed in the work of [47,48][47][48]), while MEA is required during gametophyte development and early embryogenesis [131,139][131][139]. Three PRC2 complexes have been identified in Arabidopsis defined by the presence of respective Su(z)12 homologs: EMBRYONIC FLOWER (EMF), VERNALIZATION (VRN), and FERTILIZATION INDEPENDENT SEED (FIS). The EMF and VRN complexes are associated with either CLF or SWN, and the FIS complex contains MEA or SWN as a catalytic subunit ([140[140][141][142],141,142], reviewed in the work of [3,69][3][69]). In A. thaliana, accessory subunits that can physically interact with core PRC2 subunits have been identified, including several transcription factors, such as the ASYMETRIC LEAVES (AS), TELOMERE REPEAT BINDING FACTORS (TRBs), VIVIPAROUS1/ABI3-LIKE (VAL), additional PRC-chromatin-associated enzymatic or scaffold proteins, such as LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), VERNALIZATION INSENSITIVE 3 (VIN3), VRN5, PWWP-DOMAIN INTERACTOR OF POLYCOMBS 1 (PWO1), INCURVATA11 (ICU11), components of the E3 ubiquitin ligase complexes, components of the DNA replication and chromosome segregation machineries (CTF4) or domesticated transposases (ALP1, ALP2) ([143[143][144][145][146][147][148][149],144,145,146,147,148,149], reviewed in the work of [3,33,45][3][33][45]). In spite of the well-described impact of PRC2 function on plant development, plant PRC2 composition and potential subcomplex function remain enigmatic. Additionally, structural and detailed biochemical information on plant PRC2 is not yet available, limiting detailed insight into PRC2 subunit interaction and mechanism of action in plants.
In animals, PRC2 catalyzes H3K27me1/2/3 [50], while in plants, H3K27me1 in centromeres and pericentromeres is catalyzed by the ARABIDOPSIS TRITHORAX-RELATED HMTs (ATXR5 and ATXR6) [150]. H3K27me3 is, therefore, the conserved hallmark of PRC2 enzymatic activity (Figure 3). H3K27me3 in D. melanogaster occupies broad domains that typically span more than 10 kb [151]. In mammals, H3K27me3 has two distribution patterns: large domains (>100 kb) encompassing the Hox loci and smaller domains of a few kilobases (reviewed by the authors of [75]). In addition to genic loci, H3K27me3 and PRC2 reside in poised enhancers (PEs) that often associate with bivalent genes in vertebrate pluripotent cells [152]. While PRC1 contributes to the PE marking globally [152] and also targets active enhancers in cancer cells [153], PRC2 is involved at PEs at specific loci [152]. Unlike in animals, the distribution of H3K27me3 in A. thaliana is usually limited to single genes, with the modification covering proximal parts of promoters and gene bodies with average enrichment regions of 1–1.5 kb [37,38,154,155][37][38][154][155]. Gene-limited distribution is also found in metabolic gene clusters that are organized in an operon-like manner [156]. In A. thaliana, H3K27me3 decorates approximately 20–30% of all protein-coding and miRNA genes [37,38,157][37][38][157] that are associated with plant development, hormone metabolism and response, but also with nutrient homeostasis [158[158][159],159], stress response ([160[160][161],161], reviewed in the works of [5,162,163][5][162][163]) or primary and secondary metabolism [156,161][156][161]. The potential for targeting is nevertheless more extensive, as 64% of protein-coding genes were identified as H3K27me3 targets when combining different timepoints of A. thaliana shoot apical meristem development during the transition to flowering [164].
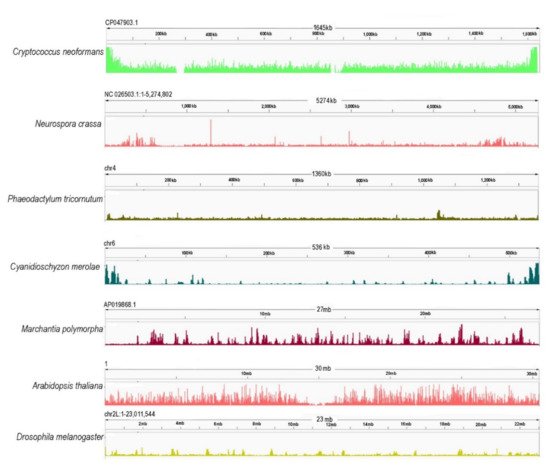
Figure 3. Chromosome-wide distribution of H3K27me3 in different model species [Cryptococcus neoformans (ASM1180120v1/PRJNA261445) [64], Neurospora crassa (NC12/PRJNA192863) [111], Phaedactylum tricornutum (ASM15095v2/PRJNA282957) [165], Cyanidioschyzon merolae (ASM9120v1/PRJNA362822) [63], Marchantia polymorpha (ASM993635v2/PRJNA553138) [166], Arabidopsis thaliana (TAIR10/PRJNA277409) [167], Drosophila melanogaster (PRJNA379297) [168]. Genome reference and accessions are given in brackets. Here, the publically available data were downloaded, cleaned by removing the library adapters, small reads (30), and low-quality (20) reads using Trim galore [169]. To map the sequenced data, Bowtie2 [170] was used with default parameters. Only mapped reads were kept with a quality threshold of 25. The filtering was performed using SAMTOOLS [171]. For the visualization of the data, IGV [172] was used, and for the parsing of the BAM files to BW files, BamCoverage—deeptools [173].
PRC2 itself lacks sequence-specific DNA-binding ability and therefore relies on accessory proteins for targeting specific loci. Several mechanisms by which PRC2 is recruited to the chromatin targets have been identified. Among these are transcription factor site-specific recruitment, interaction with RNAs, or association with chromatin features (reviewed in the work of [88]). In D. melanogaster, PRC2 is recruited to DNA targets by different transcription factors that interact with sequence motifs known as polycomb response elements (PREs) ([174[174][175],175], reviewed in the work of [176]). PREs in Drosophila are found in proximal promoter regions of developmental genes. They typically span about 1.5 kb [177] and contain numerous binding sites for a variety of DNA sequence-specific binding factors, such as the GTGT-motif-binding Combgap (Cg) [178], which together mediate PRC2 recruitment to target sites (reviewed in the work of [75]). Typical fly-like PREs are not found in mammals, and even though several mammalian PREs have been reported [179], they are not evolutionarily conserved. CpG islands, hypomethylated CG-rich chromatin regions of 1–2 kb, are associated with PRC2 recruitment in mammals, where accessory subunits rather than transcription factors mediate the recruitment ([180,181,182][180][181][182], reviewed in the work of [71]). Transcription factor-bound PRE-like elements also contribute to PRC2 recruitment in A. thaliana (reviewed in the work of [5]). Following the identification of PRC2-recruiting cis elements in the promoters of the KNOX genes [183] and the LEC2 gene [184], PRE-like elements (including GAGA motif, W-box, RY motif, GCCGCC motif, telobox motif, and others) bound by transcription factors were shown to contribute to PRC2 recruitment genome-wide [185]. Several transcription factors have been demonstrated to interact directly with PRC2 subunits to mediate the complex’s recruitment [144,146,183,186][144][146][183][186]. For instance, transcriptional repressors VIVIPAROUS1/ABI3-LIKE1 (VAL1) and VAL2 or TELOMERE REPEAT-BINDING FACTORS (TRBs) physically interact with SWN and CLF to recruit PRC2 to specific sequence motifs present in the promoters of target genes [144,146,187][144][146][187]. PRC2 recruitment through interaction with trans-acting factors that recognize cis elements is thus conserved in both plant as well as animal models (reviewed in the work of [45]).
Noncoding RNAs (ncRNAs), in particular long noncoding RNAs (lncRNAs), and nascent RNAs have also been implemented in PRC2 binding and recruitment to target sites (reviewed in the work of [188,189][188][189]). The formation of DNA-RNA hybrid structures (R-loops) is suggested to promote PRC2 recruitment in mammalian cells [190]. R-loops have been found at a variety of polycomb target gene loci, mostly found adjacent to the promoters (reviewed in the work of [191]), and interestingly, R-loop formation was detected at about one-third of PREs in Drosophila embryos [192], suggesting conservation of the mechanism. R-loops have been shown to positively and negatively impact recruitment, and their role is discussed [190,193][190][193]. lncRNAs are also known to contribute to PRC2 recruitment in A. thaliana. For instance, COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR), COOLAIR, and COLDWRAP act in repressing FLOWERING LOCUS C (FLC) transcription by H3K27me3 [194,195][194][195] and the lncRNA APOLO contributes to LHP1 recruitment, H3K27me3 enrichment, and chromatin looping at the PINOID (PID) locus that encodes a key polar auxin transport regulator [196].
Last but not least, PRC2 recruitment and activity are prevented or promoted by its interaction with the existing chromatin environment (Figure 4). Several pre-existing chromatin modifications, including H3K27ac, H3K4me3, H3K36me2/3, and cytosine methylation in CpG islands, prevent PRC2 recruitment and/or inhibit its activity ([77[77][180][197][198][199][200],180,197,198,199,200], reviewed in the work of [201]). In addition to opposing PRC2, H3K36me3 may promote H3K27me3. In mouse embryonic stem cells, Phf19, a PCL ortholog, binds to H3K36me2/H3K36me3, recruiting PRC2 and lysine demethylases to promote PRC2 activity [202]. Although biochemical evidence is limited in plants [198], H3K27me3 mainly occupies regions that are depleted for active chromatin modifications [154,155][154][155]. An exception to this is regions of bivalent chromatin where activating and repressive modifications co-localize to potentiate rapid change of gene transcription. In both plant and animals, the best well-described bivalent chromatin is marked by H3K27me3 and H3K4me3 ([155], reviewed in the work of [203]), but other active modifications may co-localize with H3K27me3 including H3K4me1 in Brassica napus [204], or H3K18ac in the camalexin biosynthesis genes in A. thaliana [205]. In mammals, H3K27me3 is promoted in a self-reinforcement loop. Pre-existing H3K27me3 is bound by EED, which allosterically stimulates PRC2 to methylate adjacent unmodified H3K27 and promote the spreading of H3K27me3 [87,206,207][87][206][207]. H3K27me3 binding activity has not been shown for the A. thaliana EED homolog FIE. Nevertheless, A. thaliana MSI1 interacts with LHP1, which binds H3K27me3, and through this may enable H3K27me3 spreading and/or post-replicative maintenance of H3K27me3 [102,208][102][208]. In addition, H2AKub, the catalytic product of PRC1, can act as a recruitment platform for PRC2 ([209[209][210],210], reviewed in the works of [45,211][45][211]). The human PRC2 cofactors JARID2 and AEBP2 bind to H2AK119ub, triggering a positive feedback loop [77] that ensures the maintenance of transcriptional repression. In animals as well as in plants, genome-wide deposition of H3K27me3 and H2Aub seem to be partially dependent on each other. In animal models (Drosophila, mouse, and human cells), most studies suggest that H2Aub modification is independent of H3K27me3 deposition, while H3K27me3 levels are decreased upon disruption of H2Aub (reviewed in the work of [212]). In A. thaliana, H3K27me3 overlaps with H2AK121ub at a subset of loci [26]. H2Aub is more widespread than H3K27me3, and its deposition is largely independent of PRC2 activity. In contrast, H3K27me3 depends on H2Aub at sites that carry both the marks, together indicating that PRC1 may be instructive for H3K27me3 in plants rather than vice versa [26]. Recently, three different chromatin states occupied either by H3K27me3 only, H2Aub only, or both modifications were described in A. thaliana, showing that accessibility increases from the inaccessible H3K27me3-only-marked chromatin to H3K27me3/H2Aub and H2Aub-only chromatin, that are mainly located at transcriptional hotspots [213]. This is in line with findings that H2Aub associates with responsive genes and its repressive function relies on H3K27me3 deposition and that H2A deubiquitination by UBP12 and UBP13 are required for stable H3K27me3-mediated repression [12,13][12][13]. Thus, while PRC1 and PRC2 activities both contribute to decreased chromatin accessibility, H3K27me3 seems to be a major contributor to stable gene repression within inaccessible chromatin regions in A. thaliana.
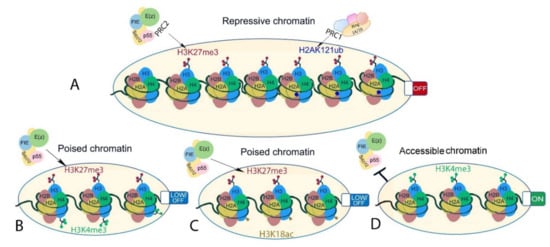
Figure 4. Different chromatin states associated or interacting with H3K27me3 in plants. (A) Facultative heterochromatin-inaccessible H3K27me3-marked chromatin (left) and inaccessible but permissive H3K27me3- and H2AK121ub-marked chromatin (right). (B,C) bivalent chromatin is marked by histone modifications with opposing effects on gene transcription. Examples are (B) H3K27me3-H3K4me3 and (C) H3K27me3 and H3K18ac. These are poised states where gene transcription is readily initiated upon stimuli. (D) Euchromatin showing chromatin state at transcriptionally active genes that is known to interfere with PRC2-mediated H3K27me3 deposition on the same histone tail in A. thaliana.
References
- Prezioso, C.; Orlando, V. Polycomb Proteins in Mammalian Cell Differentiation and Plasticity. FEBS Lett. 2011, 585, 2067–2077.
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of Gene Transcription by Polycomb Proteins. Sci. Adv. 2015, 1, e1500737.
- Mozgova, I.; Hennig, L. The Polycomb Group Protein Regulatory Network. Annu. Rev. Plant Biol. 2015.
- Yu, J.R.; Lee, C.H.; Oksuz, O.; Stafford, J.M.; Reinberg, D. PRC2 Is High Maintenance. Genes Dev. 2019, 33, 903–935.
- Shen, Q.; Lin, Y.; Li, Y.; Wang, G. Plants Dynamics of H3K27me3 Modification on Plant Adaptation to Environmental Cues. Plants 2021, 10, 1165.
- Lee, H.-G.; Kahn, T.G.; Simcox, A.; Schwartz, Y.B.; Pirrotta, V. Genome-Wide Activities of Polycomb Complexes Control Pervasive Transcription. Genome Res. 2015, 25, 1170–1181.
- Bratzel, F.; López-Torrejón, G.; Koch, M.; Del Pozo, J.C.; Calonje, M. Keeping Cell Identity in Arabidopsis Requires PRC1 RING-Finger Homologs That Catalyze H2A Monoubiquitination. Curr. Biol. 2010, 20, 1853–1859.
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H2A Ubiquitination in Polycomb Silencing. Nature 2004, 431, 873–878.
- Wiles, E.T.; Selker, E.U. H3K27 Methylation: A Promiscuous Repressive Chromatin Mark. Curr. Opin. Genet. Dev. 2017, 43, 31–37.
- Chittock, E.C.; Latwiel, S.; Miller, T.C.R.; Müller, C.W. Molecular Architecture of Polycomb Repressive Complexes. Biochem. Soc. Trans. 2017, 45, 193–205.
- Reddington, C.J.; Fellner, M.; Burgess, A.E.; Mace, P.D. Molecular Regulation of the Polycomb Repressive-Deubiquitinase. Int. J. Mol. Sci. 2020, 21, 7837.
- Derkacheva, M.; Liu, S.; Figueiredo, D.D.; Gentry, M.; Mozgova, I.; Nanni, P.; Tang, M.; Mannervik, M.; Köhler, C.; Hennig, L. H2A Deubiquitinases UBP12/13 Are Part of the Arabidopsis Polycomb Group Protein System. Nat. Plants 2016, 2, 16126.
- Kralemann, L.E.M.; Liu, S.; Trejo-Arellano, M.S.; Muñoz-Viana, R.; Köhler, C.; Hennig, L. Removal of H2Aub1 by Ubiquitin-Specific Proteases 12 and 13 Is Required for Stable Polycomb-Mediated Gene Repression in Arabidopsis. Genome Biol. 2020, 21, 144.
- Klymenko, T.; Papp, B.; Fischle, W.; Köcher, T.; Schelder, M.; Fritsch, C.; Wild, B.; Wilm, M.; Müller, J. A Polycomb Group Protein Complex with Sequence-Specific DNA-Binding and Selective Methyl-Lysine-Binding Activities. Genes Dev. 2006, 20, 1110–1122.
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57.
- Stewart-Morgan, K.R.; Petryk, N.; Groth, A. Chromatin Replication and Epigenetic Cell Memory. Nat. Cell Biol. 2020, 22, 361–371.
- Probst, A.V.; Desvoyes, B.; Gutierrez, C. Similar yet Critically Different: The Distribution, Dynamics and Function of Histone Variants. J. Exp. Bot. 2020, 71, 5191–5204.
- Hugues, A.; Jacobs, C.S.; Roudier, F. Mitotic Inheritance of PRC2-Mediated Silencing: Mechanistic Insights and Developmental Perspectives. Front. Plant Sci. 2020, 11, 262.
- Pu, L.; Sung, Z.R. PcG and TrxG in Plants—Friends or Foes. Trends Genet. 2015, 31, 252–262.
- Wang, S.; Ordonez-Rubiano, S.C.; Dhiman, A.; Jiao, G.; Strohmier, B.P.; Krusemark, C.J.; Dykhuizen, E.C. Polycomb Group Proteins in Cancer: Multifaceted Functions and Strategies for Modulation. NAR Cancer 2021, 3, zcab039.
- Dockerill, M.; Gregson, C.; O’ Donovan, D.H. Targeting PRC2 for the Treatment of Cancer: An Updated Patent Review (2016–2020). Expert Opin. Ther. Pat. 2021, 31, 119–135.
- Piunti, A.; Shilatifard, A. The Roles of Polycomb Repressive Complexes in Mammalian Development and Cancer. Nat. Rev. Mol. Cell Biol. 2021, 22, 326–345.
- Lewis, P.H. New Mutants Report. Drosoph. Inf. Serv. 1947, 21, 69.
- Sanchez-Pulido, L.; Devos, D.; Sung, Z.R.; Calonje, M. RAWUL: A New Ubiquitin-like Domain in PRC1 Ring Finger Proteins That Unveils Putative Plant and Worm PRC1 Orthologs. BMC Genom. 2008, 9, 308.
- Chen, D.; Molitor, A.; Liu, C.; Shen, W.H. The Arabidopsis PRC1-like Ring-Finger Proteins Are Necessary for Repression of Embryonic Traits during Vegetative Growth. Cell Res. 2010, 20, 1332–1344.
- Zhou, Y.; Romero-Campero, F.J.; Gómez-Zambrano, Á.; Turck, F.; Calonje, M. H2A Monoubiquitination in Arabidopsis Thaliana Is Generally Independent of LHP1 and PRC2 Activity. Genome Biol. 2017, 18, 69.
- Berke, L.; Snel, B. The Plant Polycomb Repressive Complex 1 (PRC1) Existed in the Ancestor of Seed Plants and Has a Complex Duplication History. BMC Evol. Biol. 2015, 15, 44.
- Chen, D.; Huang, Y.; Ruan, Y.; Shen, W.-H. The Evolutionary Landscape of PRC1 Core Components in Green Lineage. Planta 2016, 243, 825–846.
- Hennig, L.; Derkacheva, M. Diversity of Polycomb Group Complexes in Plants: Same Rules, Different Players? Trends Genet. 2009, 25, 414–423.
- Calonje, M. Prc1 Marks the Difference in Plant Pcg Repression. Mol. Plant 2014, 7, 459–471.
- Shaver, S.; Casas-Mollano, J.A.; Cerny, R.L.; Cerutti, H. Origin of the Polycomb Repressive Complex 2 and Gene Silencing by an E(z) Homolog in the Unicellular Alga Chlamydomonas. Epigenetics 2010, 5, 301–312.
- Sowpati, D.T.; Ramamoorthy, S.; Mishra, R.K. Expansion of the Polycomb System and Evolution of Complexity. Mech. Dev. 2015, 138, 97–112.
- Förderer, A.; Zhou, Y.; Turck, F. The Age of Multiplexity: Recruitment and Interactions of Polycomb Complexes in Plants. Curr. Opin. Plant Biol. 2016, 29, 169–178.
- van Mierlo, G.; Veenstra, G.J.C.; Vermeulen, M.; Marks, H. The Complexity of PRC2 Subcomplexes. Trends Cell Biol. 2019, 29, 660–671.
- Miller, S.A.; Damle, M.; Kim, J.; Kingston, R.E. Full Methylation of H3K27 by PRC2 Is Dispensable for Initial Embryoid Body Formation but Required to Maintain Differentiated Cell Identity. Development 2021, 148, dev196329.
- Mozgová, I.; Muñoz-Viana, R.; Hennig, L. PRC2 Represses Hormone-Induced Somatic Embryogenesis in Vegetative Tissue of Arabidopsis Thaliana. PLoS Genet. 2017, 13, e1006562.
- Turck, F.; Roudier, F.; Farrona, S.; Martin-Magniette, M.L.; Guillaume, E.; Buisine, N.; Gagnot, S.; Martienssen, R.A.; Coupland, G.; Colot, V. Arabidopsis TFL2/LHP1 Specifically Associates with Genes Marked by Trimethylation of Histone H3 Lysine 27. PLoS Genet. 2007, 3, e86.
- Zhang, X.; Clarenz, O.; Cokus, S.; Bernatavichute, Y.V.; Pellegrini, M.; Goodrich, J.; Jacobsen, S.E. Whole-Genome Analysis of Histone H3 Lysine 27 Trimethylation in Arabidopsis. PLoS Biol. 2007, 5, e129.
- Margueron, R.; Trojer, P.; Reinberg, D. The Key to Development: Interpreting the Histone Code? Curr. Opin. Genet. Dev. 2005, 15, 163–176.
- Steffen, P.A.; Ringrose, L. What Are Memories Made of? How Polycomb and Trithorax Proteins Mediate Epigenetic Memory. Nat. Rev. Mol. Cell Biol. 2014, 15, 340–356.
- Kang, S.J.; Chun, T. Structural Heterogeneity of the Mammalian Polycomb Repressor Complex in Immune Regulation. Exp. Mol. Med. 2020, 52, 1004–1015.
- Martin, C.J.; Moorehead, R.A. Polycomb Repressor Complex 2 Function in Breast Cancer (Review). Int. J. Oncol. 2020, 57, 1085–1094.
- Cao, Y.; Li, L.; Fan, Z. The Role and Mechanisms of Polycomb Repressive Complex 2 on the Regulation of Osteogenic and Neurogenic Differentiation of Stem Cells. Cell Prolif. 2021, 54, e13032.
- Guo, Y.; Zhao, S.; Wang, G.G. Polycomb Gene Silencing Mechanisms: PRC2 Chromatin Targeting, H3K27me3 “Readout”, and Phase Separation-Based Compaction. Trends Genet. 2021, 37, 547–565.
- Bieluszewski, T.; Xiao, J.; Yang, Y.; Wagner, D. PRC2 Activity, Recruitment, and Silencing: A Comparative Perspective. Trends Plant Sci. 2021, 26, 1186–1198.
- Glancy, E.; Ciferri, C.; Bracken, A.P. Structural Basis for PRC2 Engagement with Chromatin. Curr. Opin. Struct. Biol. 2021, 67, 135–144.
- Shu, J.; Chen, C.; Li, C.; Cui, Y. The Complexity of PRC2 Catalysts CLF and SWN in Plants. Biochem. Soc. Trans. 2020, 48, 2779–2789.
- Hinsch, V.; Adkins, S.; Manuela, D.; Xu, M. Post-Embryonic Phase Transitions Mediated by Polycomb Repressive Complexes in Plants. Int. J. Mol. Sci. 2021, 22, 7533.
- Déléris, A.; Berger, F.; Duharcourt, S. Role of Polycomb in the Control of Transposable Elements. Trends Genet. 2021, 37, 882–889.
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell 2002, 111, 197–208.
- Yang, Y.; Li, G. Post-Translational Modifications of PRC2: Signals Directing Its Activity. Epigenet. Chromatin 2020, 13, 47.
- Nekrasov, M.; Wild, B.; Müller, J. Nucleosome Binding and Histone Methyltransferase Activity of Drosophila PRC2. EMBO Rep. 2005, 6, 348–353.
- Wang, X.; Paucek, R.D.; Gooding, A.R.; Brown, Z.Z.; Eva, J.; Muir, T.W.; Cech, T.R. Molecular Analysis of PRC2 Recruitment to DNA in Chromatin and Its Inhibition by RNA. Nat. Struct. Mol. Biol. 2017, 24, 1028–1038.
- Wang, X.; Goodrich, K.J.; Gooding, A.R.; Youmans, D.T.; Cech, T.R.; Wang, X.; Goodrich, K.J.; Gooding, A.R.; Naeem, H.; Archer, S.; et al. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines Article Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol. Cell 2017, 65, 1056–1067.
- Kasinath, V.; Poepsel, S.; Nogales, E. Recent Structural Insights into PRC2 Regulation and Substrate Binding. Biochemistry 2019, 58, 346–354.
- Holoch, D.; Margueron, R. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem. Sci. 2017, 42, 531–542.
- Ketel, C.S.; Andersen, E.F.; Vargas, M.L.; Suh, J.; Strome, S.; Simon, J.A. Subunit Contributions to Histone Methyltransferase Activities of Fly and Worm Polycomb Group Complexes. Mol. Cell. Biol. 2005, 25, 6857–6868.
- Cao, R.; Zhang, Y. SUZ12 Is Required for Both the Histone Methyltransferase Activity and the Silencing Function of the EED-EZH2 Complex. Mol. Cell 2004, 15, 57–67.
- Pasini, D.; Bracken, A.P.; Jensen, M.R.; Denchi, E.L.; Helin, K. Suz12 Is Essential for Mouse Development and for EZH2 Histone Methyltransferase Activity. EMBO J. 2004, 23, 4061–4071.
- Kasinath, V.; Faini, M.; Poepsel, S.; Reif, D.; Feng, X.A.; Stjepanovic, G.; Aebersold, R.; Nogales, E. Structures of Human PRC2 with Its Cofactors AEBP2 and JARID2. Science 2018, 359, 940–944.
- Mosquna, A.; Katz, A.; Decker, E.L.; Rensing, S.A.; Reski, R.; Ohad, N. Regulation of Stem Cell Maintenance by the Polycomb Protein FIE Has Been Conserved during Land Plant Evolution. Development 2009, 136, 2433–2444.
- Okano, Y.; Aono, N.; Hiwatashi, Y.; Murata, T.; Nishiyama, T.; Ishikawa, T.; Kubo, M.; Hasebe, M. A Polycomb Repressive Complex 2 Gene Regulates Apogamy and Gives Evolutionary Insights into Early Land Plant Evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 16321–16326.
- Mikulski, P.; Komarynets, O.; Fachinelli, F.; Weber, A.P.M.; Schubert, D. Characterization of the Polycomb-Group Mark H3K27me3 in Unicellular Algae. Front. Plant Sci. 2017, 8, 607.
- Dumesic, P.A.; Homer, C.M.; Moresco, J.J.; Pack, L.R.; Shanle, E.K.; Coyle, S.M.; Strahl, B.D.; Fujimori, D.G.; Yates, J.R.; Madhani, H.D. Product Binding Enforces the Genomic Specificity of a Yeast Polycomb Repressive Complex. Cell 2015, 160, 204–218.
- Zhao, X.; Rastogi, A.; Deton Cabanillas, A.F.; Ait Mohamed, O.; Cantrel, C.; Lombard, B.; Murik, O.; Genovesio, A.; Bowler, C.; Bouyer, D.; et al. Genome Wide Natural Variation of H3K27me3 Selectively Marks Genes Predicted to Be Important for Cell Differentiation in Phaeodactylum Tricornutum. New Phytol. 2021, 229, 3208–3220.
- Gall Trošelj, K.; Novak Kujundzic, R.; Ugarkovic, D. Polycomb Repressive Complex’s Evolutionary Conserved Function: The Role of EZH2 Status and Cellular Background. Clin. Epigenet. 2016, 8, 55.
- Deevy, O.; Bracken, A.P. PRC2 Functions in Development and Congenital Disorders. Development 2019, 146, dev181354.
- Lewis, Z.A. Polycomb Group Systems in Fungi: New Models for Understanding Polycomb Repressive Complex 2. Trends Genet. 2017, 33, 220–231.
- Derkacheva, M.; Hennig, L. Variations on a Theme: Polycomb Group Proteins in Plants. J. Exp. Bot. 2014, 65, 2769–2784.
- Smits, A.H.; Jansen, P.W.T.C.; Poser, I.; Hyman, A.A.; Vermeulen, M. Stoichiometry of Chromatin-Associated Protein Complexes Revealed by Label-Free Quantitative Mass Spectrometry-Based Proteomics. Nucleic Acids Res. 2013, 41, e28.
- Laugesen, A.; Højfeldt, J.W.; Helin, K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell 2019, 74, 8–18.
- Huang, Y.; Su, T.; Wang, C.; Dong, L.; Liu, S.; Zhu, Y.; Hao, K.; Xia, Y.; Jiang, Q.; Qin, J. Rbbp4 Suppresses Premature Differentiation of Embryonic Stem Cells. Stem Cell Rep. 2021, 16, 566–581.
- Xu, C.; Min, J. Structure and Function of WD40 Domain Proteins. Protein Cell 2011, 2, 202–214.
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595.
- Margueron, R.; Reinberg, D. The Polycomb Complex PRC2 and Its Mark in Life. Nature 2011, 469, 343–349.
- Giner-Laguarda, N.; Vidal, M. Functions of Polycomb Proteins on Active Targets. Epigenomes 2020, 4, 17.
- Kasinath, V.; Beck, C.; Sauer, P.; Poepsel, S.; Kosmatka, J.; Faini, M.; Toso, D.; Aebersold, R.; Nogales, E. JARID2 and AEBP2 Regulate PRC2 in the Presence of H2AK119ub1 and Other Histone Modifications. Science 2021, 371, eabc3393.
- Han, Z.; Xing, X.; Hu, M.; Zhang, Y.; Liu, P.; Chai, J. Structural Basis of EZH2 Recognition by EED. Structure 2007, 15, 1306–1315.
- Jiao, L.; Liu, X. Structural Basis of Histone H3K27 Trimethylation by an Active Polycomb Repressive Complex 2. Science 2015, 350, aac4383.
- Bratkowski, M.; Yang, X.; Liu, X. Polycomb Repressive Complex 2 in an Autoinhibited State. J. Biol. Chem. 2017, 292, 13323–13332.
- Uckelmann, M.; Davidovich, C. Not Just a Writer: PRC2 as a Chromatin Reader. Biochem. Soc. Trans. 2021, 49, 1159–1170.
- Shi, Y.; Wang, X.X.; Zhuang, Y.W.; Jiang, Y.; Melcher, K.; Xu, H.E. Structure of the PRC2 Complex and Application to Drug Discovery. Acta Pharmacol. Sin. 2017, 38, 963–976.
- Moritz, L.E.; Trievel, R.C. Structure, Mechanism, and Regulation of Polycomb-Repressive Complex 2. J. Biol. Chem. 2018, 293, 13805–13814.
- Chammas, P.; Mocavini, I.; Di Croce, L. Engaging Chromatin: PRC2 Structure Meets Function. Br. J. Cancer 2020, 122, 315–328.
- The PyMOL Molecular Graphics System; Version 2.4.1; Schrödinger, LLC.: New York, NY, USA, 2020.
- Kasinath, V.; Nogales, E.; Beck, C.; Sauer, P.; Poepsel, S.; Kosmatka, J.; Faini, M.; Toso, D.; Aebersold, R. PRC2-AEBP2-JARID2 Bound to H2AK119ub1 Nucleosome. Available online: https://www.wwpdb.org/pdb?id=pdb_00006wkr (accessed on 20 November 2021).
- Li, L.; Zhang, H.; Zhang, M.; Zhao, M.; Feng, L.; Luo, X.; Gao, Z.; Huang, Y.; Ardayfio, O.; Zhang, J.H.; et al. Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED. PLoS ONE 2017, 12, e0169855.
- Blackledge, N.P.; Klose, R.J. The Molecular Principles of Gene Regulation by Polycomb Repressive Complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 815–833.
- Huang, Y.; Chen, D.H.; Liu, B.Y.; Shen, W.H.; Ruan, Y. Conservation and Diversification of Polycomb Repressive Complex 2 (PRC2) Proteins in the Green Lineage. Brief. Funct. Genom. 2017, 16, 106–119.
- Chen, L.J.; Diao, Z.Y.; Specht, C.; Sung, Z.R. Molecular Evolution of VEF-Domain-Containing PcG Genes in Plants. Mol. Plant 2009, 2, 738–754.
- Luo, M.; Platten, D.; Chaudhury, A.; Peacock, W.J.; Dennis, E.S. Expression, Imprinting, and Evolution of Rice Homologs of the Polycomb Group Genes. Mol. Plant 2009, 2, 711–723.
- Nakamura, M.; Batista, R.A.; Köhler, C.; Hennig, L. Polycomb Repressive Complex 2-Mediated Histone Modification H3K27me3 Is Associated with Embryogenic Potential in Norway Spruce. J. Exp. Bot. 2020, 71, 6366–6378.
- Liu, X.; Zhou, C.; Zhao, Y.; Zhou, S.; Wang, W.; Zhou, D.X. The Rice Enhancer of Zeste Genes SDG711 and SDG718 Are Respectively Involved in Long Day and Short Day Signaling to Mediate the Accurate Photoperiod Control of Flowering Time. Front. Plant Sci. 2014, 5, 591.
- Ni, J.; Ma, X.; Feng, Y.; Tian, Q.; Wang, Y.; Xu, N.; Tang, J.; Wang, G. Updating and Interaction of Polycomb Repressive Complex 2 Components in Maize (Zea Mays). Planta 2019, 250, 573–588.
- Cheng, X.; Pan, M.; Zhiguo, E.; Zhou, Y.; Niu, B.; Chen, C. The Maternally Expressed Polycomb Group Gene OsEMF2a Is Essential for Endosperm Cellularization and Imprinting in Rice. Plant Commun. 2020, 2, 100092.
- Conrad, L.J.; Khanday, I.; Johnson, C.; Guiderdoni, E.; An, G.; Vijayraghavan, U.; Sundaresan, V. The Polycomb Group Gene EMF2B Is Essential for Maintenance of Floral Meristem Determinacy in Rice. Plant J. 2014, 80, 883–894.
- Strejčková, B.; Čegan, R.; Pecinka, A.; Milec, Z.; Šafář, J. Identification of Polycomb Repressive Complex 1 and 2 Core Components in Hexaploid Bread Wheat. BMC Plant Biol. 2020, 20, 175.
- Higgins, J.A.; Bailey, P.C.; Laurie, D.A. Comparative Genomics of Flowering Time Pathways Using Brachypodium Distachyon as a Model for the Temperate Grasses. PLoS ONE 2010, 5, e10065.
- Lomax, A.; Woods, D.P.; Dong, Y.; Bouché, F.; Rong, Y.; Mayer, K.S.; Zhong, X.; Amasino, R.M. An Ortholog of CURLY LEAF/ENHANCER OF ZESTE like-1 Is Required for Proper Flowering in Brachypodium Distachyon. Plant J. 2018, 93, 871–882.
- Kapazoglou, A.; Tondelli, A.; Papaefthimiou, D.; Ampatzidou, H.; Francia, E.; Stanca, M.A.; Bladenopoulos, K.; Tsaftaris, A.S. Epigenetic Chromatin Modifiers in Barley: IV. The Study of Barley Polycomb Group (PcG) Genes during Seed Development and in Response to External ABA. BMC Plant Biol. 2010, 10, 73.
- Tonosaki, K.; Kinoshita, T. Possible Roles for Polycomb Repressive Complex 2 in Cereal Endosperm. Front. Plant Sci. 2015, 6, 144.
- Derkacheva, M.; Steinbach, Y.; Wildhaber, T.; Mozgová, I.; Mahrez, W.; Nanni, P.; Bischof, S.; Gruissem, W.; Hennig, L. Arabidopsis MSI1 Connects LHP1 to PRC2 Complexes. EMBO J. 2013, 32, 2073–2085.
- Zhao, X.; Deton Cabanillas, A.F.; Veluchamy, A.; Bowler, C.; Vieira, F.R.J.; Tirichine, L. Probing the Diversity of Polycomb and Trithorax Proteins in Cultured and Environmentally Sampled Microalgae. Front. Mar. Sci. 2020, 7, 189.
- Bourdareau, S.; Tirichine, L.; Lombard, B.; Loew, D.; Scornet, D.; Wu, Y.; Coelho, S.M.; Cock, J.M. Histone Modifications during the Life Cycle of the Brown Alga Ectocarpus. Genome Biol. 2021, 22, 12.
- Rzeszutek, I.; Maurer-Alcalá, X.X.; Nowacki, M. Programmed Genome Rearrangements in Ciliates. Cell. Mol. Life Sci. 2020, 77, 4615–4629.
- Frapporti, A.; Miró Pina, C.; Arnaiz, O.; Holoch, D.; Kawaguchi, T.; Humbert, A.; Eleftheriou, E.; Lombard, B.; Loew, D.; Sperling, L.; et al. The Polycomb Protein Ezl1 Mediates H3K9 and H3K27 Methylation to Repress Transposable Elements in Paramecium. Nat. Commun. 2019, 10, 2710.
- Xu, J.; Zhao, X.; Mao, F.; Basrur, V.; Ueberheide, B.; Chait, B.T.; David Allis, C.; Taverna, S.D.; Gao, S.; Wang, W.; et al. A Polycomb Repressive Complex Is Required for RNAi-Mediated Heterochromatin Formation and Dynamic Distribution of Nuclear Bodies. Nucleic Acids Res. 2021, 49, 5407–5425.
- Nabeel-Shah, S.; Garg, J.; Saettone, A.; Ashraf, K.; Lee, H.; Wahab, S.; Ahmed, N.; Fine, J.; Derynck, J.; Pu, S.; et al. Functional Characterization of RebL1 Highlights the Evolutionary Conservation of Oncogenic Activities of the RBBP4/7 Orthologue in Tetrahymena Thermophila. Nucleic Acids Res. 2021, 49, 6196–6212.
- Ridenour, J.B.; Möller, M.; Freitag, M. Polycomb Repression without Bristles: Facultative Heterochromatin and Genome Stability in Fungi. Genes 2020, 11, 638.
- Connolly, L.R.; Smith, K.M.; Freitag, M. The Fusarium Graminearum Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters. PLoS Genet. 2013, 9, e1003916.
- Jamieson, K.; Rountree, M.R.; Lewis, Z.A.; Stajich, J.E.; Selker, E.U. Regional Control of Histone H3 Lysine 27 Methylation in Neurospora. Proc. Natl. Acad. Sci. USA 2013, 110, 6027–6032.
- McNaught, K.J.; Wiles, E.T.; Selker, E.U. Identification of a PRC2 Accessory Subunit Required for Subtelomeric H3K27 Methylation in Neurospora Crassa. Mol. Cell. Biol. 2020, 40, e00003-20.
- Sebé-Pedrós, A.; Ballaré, C.; Parra-Acero, H.; Chiva, C.; Tena, J.J.; Sabidó, E.; Gómez-Skarmeta, J.L.; Di Croce, L.; Ruiz-Trillo, I. The Dynamic Regulatory Genome of Capsaspora and the Origin of Animal Multicellularity. Cell 2016, 165, 1224–1237.
- Gaiti, F.; Jindrich, K.; Fernandez-Valverde, S.L.; Roper, K.E.; Degnan, B.M.; Tanurdžić, M. Landscape of Histone Modifications in a Sponge Reveals the Origin of Animal Cis-Regulatory Complexity. eLife 2017, 6, e22194.
- Genikhovich, G.; Kürn, U.; Hemmrich, G.; Bosch, T.C.G. Discovery of Genes Expressed in Hydra Embryogenesis. Dev. Biol. 2006, 289, 466–481.
- Pillai, A.; Gungi, A.; Reddy, P.C.; Galande, S. Epigenetic Regulation in Hydra: Conserved and Divergent Roles. Front. Cell Dev. Biol. 2021, 9, 663208.
- PREDICTED: Polycomb Protein Suz12-like . Available online: Https://Www.Ncbi.Nlm.Nih.Gov/Protein/XP_012556196.1?Report=genbank&log$=prottop&blast_rank=1&RID=R2KAB9PA013 (accessed on 21 October 2021).
- PREDICTED: Histone-Binding Protein RBBP7 . Available online: Https://Www.Ncbi.Nlm.Nih.Gov/Protein/XP_012554465.1?Report=genbank&log$=prottop&blast_rank=1&RID=R2KUGECN013 (accessed on 21 October 2021).
- Herz, H.-M.; Mohan, M.; Garrett, A.S.; Miller, C.; Casto, D.; Zhang, Y.; Seidel, C.; Haug, J.S.; Florens, L.; Washburn, M.P.; et al. Polycomb Repressive Complex 2-Dependent and -Independent Functions of Jarid2 in Transcriptional Regulation in Drosophila. Mol. Cell. Biol. 2012, 32, 1683–1693.
- Tolhuis, B.; Muijrers, I.; de Wit, E.; Teunissen, H.; Talhout, W.; van Steensel, B.; van Lohuizen, M. Genome-Wide Profiling of PRC1 and PRC2 Polycomb Chromatin Binding in Drosophila Melanogaster. Nat. Genet. 2006, 38, 694–699.
- Siebold, A.P.; Banerjee, R.; Tie, F.; Kiss, D.L.; Moskowitz, J.; Harte, P.J. Polycomb Repressive Complex 2 and Trithorax Modulate Drosophila Longevity and Stress Resistance. Proc. Natl. Acad. Sci. USA 2010, 107, 169.
- Bender, L.B.; Cao, R.; Zhang, Y.; Strome, S. The MES-2/MES-3/MES-6 Complex and Regulation of Histone H3 Methylation in C. Elegans. Curr. Biol. 2004, 14, 1639–1643.
- Gaydos, L.J.; Wang, W.; Strome, S. H3K27me and PRC2 Transmit a Memory of Repression across Generations and during Development. Science 2014, 345, 1515–1518.
- Brooun, A.; Gajiwala, K.S.; Deng, Y.L.; Liu, W.; Bolaños, B.; Bingham, P.; He, Y.A.; Diehl, W.; Grable, N.; Kung, P.P.; et al. Polycomb Repressive Complex 2 Structure with Inhibitor Reveals a Mechanism of Activation and Drug Resistance. Nat. Commun. 2016, 7, 11384.
- PREDICTED: LOW QUALITY PROTEIN: Polycomb Protein SUZ12 . Available online: Https://Www.Ncbi.Nlm.Nih.Gov/Protein/XP_008112590.1?Report=genbank&log$=prottop&blast_rank=1&RID=R2PMHJ83016 (accessed on 21 October 2021).
- PREDICTED: Polycomb Protein EED . Available online: Https://Www.Ncbi.Nlm.Nih.Gov/Protein/XP_003219406.1?Report=genbank&log$=prottop&blast_rank=1&RID=R2PDKH5B013 (accessed on 21 October 2021).
- PREDICTED: Histone-Binding Protein RBBP4 Isoform X2 . Available online: Https://Www.Ncbi.Nlm.Nih.Gov/Protein/XP_003230003.3?Report=genbank&log$=prottop&blast_rank=1&RID=R2P2UE3N013 (accessed on 21 October 2021).
- Grossniklaus, U.; Paro, R. Transcriptional Silencing by Polycomb Group Proteins Cellular Memory. Cold Spring Harb. Perspect. Biol. 2014, 6, a019331.
- He, A.; Ma, Q.; Cao, J.; von Gise, A.; Zhou, P.; Xie, H.; Zhang, B.; Hsing, M.; Christodoulou, D.C.; Cahan, P.; et al. Polycomb Repressive Complex 2 Regulates Normal Development of the Mouse Heart. Circ. Res. 2012, 110, 406–415.
- Hennig, L.; Taranto, P.; Walser, M.; Schönrock, N.; Gruissem, W. Arabidopsis MSI1 Is Required for Epigenetic Maintenance of Reproductive Development. Development 2003, 130, 2555–2565.
- Köhler, C.; Hennig, L.; Bouveret, R.; Gheyselinck, J.; Grossniklaus, U.; Gruissem, W. Arabidopsis MSI1 Is a Component of the MEA/FIE Polycomb Group Complex and Required for Seed Development. EMBO J. 2003, 22, 4804–4814.
- Hennig, L.; Bouveret, R.; Gruissem, W. MSI1-like Proteins: An Escort Service for Chromatin Assembly and Remodeling Complexes. Trends Cell Biol. 2005, 15, 295–302.
- Doyle, M.R.; Amasino, R.M. A Single Amino Acid Change in the Enhancer of Zeste Ortholog CURLY LEAF Results in Vernalization-Independent, Rapid Flowering in Arabidopsis. Plant Physiol. 2009, 151, 1688–1697.
- Xu, M.; Hu, T.; Smith, M.R.; Poethig, R.S. Epigenetic Regulation of Vegetative Phase Change in Arabidopsis. Plant Cell 2016, 28, 28–41.
- Schubert, D.; Primavesi, L.; Bishopp, A.; Roberts, G.; Doonan, J.; Jenuwein, T.; Goodrich, J. Silencing by Plant Polycomb-Group Genes Requires Dispersed Trimethylation of Histone H3 at Lysine 27. EMBO J. 2006, 25, 4638–4649.
- Chanvivattana, Y.; Bishopp, A.; Schubert, D.; Stock, C.; Moon, Y.H.; Sung, Z.R.; Goodrich, J. Interaction of Polycomb-Group Proteins Controlling Flowering in Arabidopsis. Development 2004, 131, 5263–5276.
- Aichinger, E.; Villar, C.B.R.; Farrona, S.; Reyes, J.C.; Hennig, L.; Köhler, C. CHD3 Proteins and Polycomb Group Proteins Antagonistically Determine Cell Identity in Arabidopsis. PLoS Genet. 2009, 5, e1000605.
- Aichinger, E.; Villar, C.B.R.; Di Mambro, R.; Sabatini, S.; Köhler, C. The CHD3 Chromatin Remodeler PICKLE and Polycomb Group Proteins Antagonistically Regulate Meristem Activity in the Arabidopsis Root. Plant Cell 2011, 23, 1047–1060.
- Simonini, S.; Bemer, M.; Bencivenga, S.; Gagliardini, V.; Pires, N.D.; Desvoyes, B.; van der Graaff, E.; Gutierrez, C.; Grossniklaus, U. The Polycomb Group Protein MEDEA Controls Cell Proliferation and Embryonic Patterning in Arabidopsis. Dev. Cell 2021, 56, 1945–1960.e7.
- Moon, Y.H.; Chen, L.; Pan, R.L.; Chang, H.S.; Zhu, T.; Maffeo, D.M.; Sung, Z.R. Erratum: EMF Genes Maintain Vegetative Development by Repressing the Flower Program in Arabidopsis. Plant Cell 2003, 15, 681–693.
- De Lucia, F.; Crevillen, P.; Jones, A.M.E.; Greb, T.; Dean, C. A PHD-Polycomb Repressive Complex 2 Triggers the Epigenetic Silencing of FLC during Vernalization. Proc. Natl. Acad. Sci. USA 2008, 105, 16831–16836.
- Zhang, S.; Wang, D.; Zhang, H.; Skaggs, M.I.; Lloyd, A.; Ran, D.; An, L.; Schumaker, K.S.; Drews, G.N.; Yadegari, R. FERTILIZATION-INDEPENDENT SEED-Polycomb Repressive Complex 2 Plays a Dual Role in Regulating Type i MADS-Box Genes in Early Endosperm Development. Plant Physiol. 2018, 177, 285–299.
- Bloomer, R.H.; Hutchison, C.E.; Bäurle, I.; Walker, J.; Fang, X.; Perera, P.; Velanis, C.N.; Gümüs, S.; Spanos, C.; Rappsilber, J.; et al. The Arabidopsis Epigenetic Regulator ICU11 as an Accessory Protein of Polycomb Repressive Complex 2. Proc. Natl. Acad. Sci. USA 2020, 117, 16660.
- Zhou, Y.; Wang, Y.; Krause, K.; Yang, T.; Dongus, J.A.; Zhang, Y.; Turck, F. Telobox Motifs Recruit CLF/SWN-PRC2 for H3K27me3 Deposition via TRB Factors in Arabidopsis. Nat. Genet. 2018, 50, 638–644.
- Hohenstatt, M.L.; Mikulski, P.; Komarynets, O.; Klose, C.; Kycia, I.; Jeltsch, A.; Farrona, S.; Schubert, D. PWWP-DOMAIN INTERACTOR OF POLYCOMBS1 Interacts with Polycomb-Group Proteins and Histones and Regulates Arabidopsis Flowering and Development. Plant Cell 2018, 30, 117–133.
- Yuan, L.; Song, X.; Zhang, L.; Yu, Y.; Liang, Z.; Lei, Y.; Ruan, J.; Tan, B.; Liu, J.; Li, C. The Transcriptional Repressors VAL1 and VAL2 Recruit PRC2 for Genome-Wide Polycomb Silencing in Arabidopsis. Nucleic Acids Res. 2021, 49, 98–113.
- Zhang, P.; Zhu, C.; Geng, Y.; Wang, Y.; Yang, Y.; Liu, Q.; Guo, W.; Chachar, S.; Riaz, A.; Yan, S.; et al. Rice and Arabidopsis Homologs of Yeast CHROMOSOME TRANSMISSION FIDELITY PROTEIN 4 Commonly Interact with Polycomb Complexes but Exert Divergent Regulatory Functions. Plant Cell 2021, 33, 1417–1429.
- Velanis, C.N.; Perera, P.; Thomson, B.; de Leau, E.; Liang, S.C.; Hartwig, B.; Förderer, A.; Thornton, H.; Arede, P.; Chen, J.; et al. The Domesticated Transposase ALP2 Mediates Formation of a Novel Polycomb Protein Complex by Direct Interaction with MSI1, a Core Subunit of Polycomb Repressive Complex 2 (PRC2). PLoS Genet. 2020, 16, e1008681.
- Zhou, Y.; Tergemina, E.; Cui, H.; Förderer, A.; Hartwig, B.; Velikkakam James, G.; Schneeberger, K.; Turck, F. Ctf4-Related Protein Recruits LHP1-PRC2 to Maintain H3K27me3 Levels in Dividing Cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 4833.
- Jacob, Y.; Feng, S.; LeBlanc, C.A.; Bernatavichute, Y.V.; Stroud, H.; Cokus, S.; Johnson, L.M.; Pellegrini, M.; Jacobsen, S.E.; Michaels, S.D. ATXR5 and ATXR6 Are H3K27 Monomethyltransferases Required for Chromatin Structure and Gene Silencing. Nat. Struct. Mol. Biol. 2009, 16, 763–768.
- Schwartz, Y.B.; Kahn, T.G.; Nix, D.A.; Li, X.-Y.; Bourgon, R.; Biggin, M.; Pirrotta, V. Genome-Wide Analysis of Polycomb Targets in Drosophila Melanogaster. Nat. Genet. 2006, 38, 700–705.
- Crispatzu, G.; Rehimi, R.; Pachano, T.; Bleckwehl, T.; Cruz-Molina, S.; Xiao, C.; Mahabir, E.; Bazzi, H.; Rada-Iglesias, A. The Chromatin, Topological and Regulatory Properties of Pluripotency-Associated Poised Enhancers Are Conserved in Vivo. Nat. Commun. 2021, 12, 4344.
- Chan, H.L.; Beckedorff, F.; Zhang, Y.; Garcia-Huidobro, J.; Jiang, H.; Colaprico, A.; Bilbao, D.; Figueroa, M.E.; LaCava, J.; Shiekhattar, R.; et al. Polycomb Complexes Associate with Enhancers and Promote Oncogenic Transcriptional Programs in Cancer through Multiple Mechanisms. Nat. Commun. 2018, 9, 3377.
- Roudier, F.; Ahmed, I.; Bérard, C.; Sarazin, A.; Mary-Huard, T.; Cortijo, S.; Bouyer, D.; Caillieux, E.; Duvernois-Berthet, E.; Al-Shikhley, L.; et al. Integrative Epigenomic Mapping Defines Four Main Chromatin States in Arabidopsis. EMBO J. 2011, 30, 1928–1938.
- Sequeira-Mendes, J.; Aragüez, I.; Peiró, R.; Mendez-Giraldez, R.; Zhang, X.; Jacobsen, S.E.; Bastolla, U.; Gutierrez, C. The Functional Topography of the Arabidopsis Genome Is Organized in a Reduced Number of Linear Motifs of Chromatin States. Plant Cell 2014, 26, 2351–2366.
- Yu, N.; Nützmann, H.W.; Macdonald, J.T.; Moore, B.; Field, B.; Berriri, S.; Trick, M.; Rosser, S.J.; Kumar, S.V.; Freemont, P.S.; et al. Delineation of Metabolic Gene Clusters in Plant Genomes by Chromatin Signatures. Nucleic Acids Res. 2016, 44, 2255–2265.
- Lafos, M.; Kroll, P.; Hohenstatt, M.L.; Thorpe, F.L.; Clarenz, O.; Schubert, D. Dynamic Regulation of H3K27 Trimethylation during Arabidopsis Differentiation. PLoS Genet. 2011, 7, e1002040.
- Bellegarde, F.; Herbert, L.; Séré, D.; Caillieux, E.; Boucherez, J.; Fizames, C.; Roudier, F.; Gojon, A.; Martin, A. Polycomb Repressive Complex 2 Attenuates the Very High Expression of the Arabidopsis Gene NRT2.1. Sci. Rep. 2018, 8, 7905.
- Park, E.Y.; Tsuyuki, K.M.; Hu, F.; Lee, J.; Jeong, J. PRC2-Mediated H3K27me3 Contributes to Transcriptional Regulation of FIT-Dependent Iron Deficiency Response. Front. Plant Sci. 2019, 10, 627.
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic Priming of Arabidopsis Seedlings Establishes a Long-Term Somatic Memory Accompanied by Specific Changes of the Epigenome. Genome Biol. 2013, 14, R59.
- Chica, C.; Louis, A.; Roest Crollius, H.; Colot, V.; Roudier, F. Comparative Epigenomics in the Brassicaceae Reveals Two Evolutionarily Conserved Modes of PRC2-Mediated Gene Regulation. Genome Biol. 2017, 18, 207.
- Lämke, J.; Bäurle, I. Epigenetic and Chromatin-Based Mechanisms in Environmental Stress Adaptation and Stress Memory in Plants. Genome Biol. 2017, 18, 124.
- Kim, J.-M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin Changes in Response to Drought, Salinity, Heat, and Cold Stresses in Plants. Front. Plant Sci. 2015, 6, 114.
- You, Y.; Sawikowska, A.; Neumann, M.; Posé, D.; Capovilla, G.; Langenecker, T.; Neher, R.A.; Krajewski, P.; Schmid, M. Temporal Dynamics of Gene Expression and Histone Marks at the Arabidopsis Shoot Meristem during Flowering. Nat. Commun. 2017, 8, 207.
- Veluchamy, A.; Rastogi, A.; Lin, X.; Lombard, B.; Murik, O.; Thomas, Y.; Dingli, F.; Rivarola, M.; Ott, S.; Liu, X.; et al. An Integrative Analysis of Post-Translational Histone Modifications in the Marine Diatom Phaeodactylum Tricornutum. Genome Biol. 2015, 16, 102.
- Montgomery, S.A.; Tanizawa, Y.; Galik, B.; Wang, N.; Ito, T.; Mochizuki, T.; Akimcheva, S.; Bowman, J.L.; Cognat, V.; Maréchal-Drouard, L.; et al. Chromatin Organization in Early Land Plants Reveals an Ancestral Association between H3K27me3, Transposons, and Constitutive Heterochromatin. Curr. Biol. 2020, 30, 573–588.e7.
- Moreno-Romero, J.; Jiang, H.; Santos-González, J.; Köhler, C. Parental Epigenetic Asymmetry of PRC2-Mediated Histone Modifications in the Arabidopsis Endosperm. EMBO J. 2016, 35, 1298–1311.
- Ma, Z.; Wang, H.; Cai, Y.; Wang, H.; Niu, K.; Wu, X.; Ma, H.; Yang, Y.; Tong, W.; Liu, F.; et al. Epigenetic Drift of H3K27me3 in Aging Links Glycolysis to Healthy Longevity in Drosophila. Elife 2018, 7, e35368.
- Trim Galore. Available online: http://www.Bioinformatics.Babraham.Ac.Uk/Projects/Trim_galore/ (accessed on 7 January 2022).
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359.
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup the Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079.
- Robinson, J.T.; Thorvaldsdóttir, H.; Wenger, A.M.; Zehir, A.; Mesirov, J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017, 77, e31–e34.
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. DeepTools2: A next Generation Web Server for Deep-Sequencing Data Analysis. Nucleic Acids Res. 2016, 44, W160–W165.
- Bredesen, B.A.; Rehmsmeier, M. DNA Sequence Models of Genome-Wide Drosophila Melanogaster Polycomb Binding Sites Improve Generalization to Independent Polycomb Response Elements. Nucleic Acids Res. 2019, 47, 7781–7797.
- Erokhin, M.; Gorbenko, F.; Lomaev, D.; Mazina, M.Y.; Mikhailova, A.; Garaev, A.K.; Parshikov, A.; Vorobyeva, N.E.; Georgiev, P.; Schedl, P.; et al. Boundaries Potentiate Polycomb Response Element-Mediated Silencing. BMC Biol. 2021, 19, 113.
- Kassis, J.A.; Brown, J.L. Polycomb Group Response Elements in Drosophila and Vertebrates. Adv. Genet. 2013, 81, 83–118.
- Schorderet, P.; Lonfat, N.; Darbellay, F.; Tschopp, P.; Gitto, S.; Soshnikova, N.; Duboule, D. A Genetic Approach to the Recruitment of PRC2 at the HoxD Locus. PLoS Genet. 2013, 9, e1003951.
- Ray, P.; De, S.; Mitra, A.; Bezstarosti, K.; Demmers, J.A.A.; Pfeifer, K.; Kassis, J.A. Combgap Contributes to Recruitment of Polycomb Group Proteins in Drosophila. Proc. Natl. Acad. Sci. USA 2016, 113, 3826–3831.
- Du, J.; Kirk, B.; Zeng, J.; Ma, J.; Wang, Q. Three Classes of Response Elements for Human PRC2 and MLL1/2-Trithorax Complexes. Nucleic Acids Res. 2018, 46, 8848–8864.
- Lynch, M.D.; Smith, A.J.H.; De Gobbi, M.; Flenley, M.; Hughes, J.R.; Vernimmen, D.; Ayyub, H.; Sharpe, J.A.; Sloane-Stanley, J.A.; Sutherland, L.; et al. An Interspecies Analysis Reveals a Key Role for Unmethylated CpG Dinucleotides in Vertebrate Polycomb Complex Recruitment. EMBO J. 2012, 31, 317–329.
- Chen, S.; Jiao, L.; Liu, X.; Yang, X.; Liu, X. A Dimeric Structural Scaffold for PRC2-PCL Targeting to CpG Island Chromatin. Mol. Cell 2020, 77, 1265–1278.
- Mendenhall, E.M.; Koche, R.P.; Truong, T.; Zhou, V.W.; Issac, B.; Chi, A.S.; Ku, M.; Bernstein, B.E. GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells. PLoS Genet. 2010, 6, e1001244.
- Lodha, M.; Marco, C.F.; Timmermans, M.C.P. The ASYMMETRIC LEAVES Complex Maintains Repression of KNOX Homeobox Genes via Direct Recruitment of Polycomb-Repressive Complex2. Genes Dev. 2013, 27, 596–601.
- Berger, N.; Dubreucq, B.; Roudier, F.; Dubos, C.; Lepiniec, L. Transcriptional Regulation of Arabidopsis LEAFY COTYLEDON2 Involves RLE, a Cis-Element That Regulates Trimethylation of Histone H3 at Lysine-27. Plant Cell 2011, 23, 4065–4078.
- Xiao, J.; Jin, R.; Yu, X.; Shen, M.; Wagner, J.D.; Pai, A.; Song, C.; Zhuang, M.; Klasfeld, S.; He, C.; et al. Cis and Trans Determinants of Epigenetic Silencing by Polycomb Repressive Complex 2 in Arabidopsis. Nat. Genet. 2017, 49, 1546–1552.
- Mu, Y.; Zou, M.; Sun, X.; He, B.; Xu, X.; Liu, Y.; Zhang, L.; Chi, W. BASIC PENTACYSTEINE Proteins Repress Abscisic Acid INSENSITIVE 4 Expression via Direct Recruitment of the Polycomb-Repressive Complex 2 in Arabidopsis Root Development. Plant Cell Physiol. 2017, 58, 607–621.
- Fouracre, J.P.; He, J.; Chen, V.J.; Sidoli, S.; Scott Poethig, R. VAL Genes Regulate Vegetative Phase Change via MiR156-Dependent and Independent Mechanisms. PLoS Genet. 2021, 17, e1009626.
- Brockdorff, N. Noncoding RNA and Polycomb Recruitment. RNA 2013, 19, 429–442.
- Davidovich, C.; Cech, T.R. The Recruitment of Chromatin Modifiers by Long Noncoding RNAs: Lessons from PRC2. Rna 2015, 21, 2007–2022.
- Skourti-Stathaki, K.; Torlai Triglia, E.; Warburton, M.; Voigt, P.; Bird, A.; Pombo, A. R-Loops Enhance Polycomb Repression at a Subset of Developmental Regulator Genes. Mol. Cell 2019, 73, 930–945.
- Allison, D.F.; Wang, G.G. R-Loops: Formation, Function, and Relevance to Cell Stress. Cell Stress 2019, 3, 38–46.
- Alecki, C.; Chiwara, V.; Sanz, L.A.; Grau, D.; Arias Pérez, O.; Boulier, E.L.; Armache, K.J.; Chédin, F.; Francis, N.J. RNA-DNA Strand Exchange by the Drosophila Polycomb Complex PRC2. Nat. Commun. 2020, 11, 1781.
- Chen, P.B.; Chen, H.V.; Acharya, D.; Rando, O.J.; Fazzio, T.G. R Loops Regulate Promoter-Proximal Chromatin Architecture and Cellular Differentiation. Nat. Struct. Mol. Biol. 2015, 22, 999–1007.
- Tian, Y.; Zheng, H.; Zhang, F.; Wang, S.; Ji, X.; Xu, C.; He, Y.; Ding, Y. PRC2 Recruitment and H3K27me3 Deposition at FLC Require FCA Binding of COOLAIR. Sci. Adv. 2019, 5, eaau7246.
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR Mediates the Coordinated Switching of Chromatin States at FLC during Vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165.
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding Transcription by Alternative RNA Polymerases Dynamically Regulates an Auxin-Driven Chromatin Loop. Mol. Cell 2014, 55, 383–396.
- Cai, L.; Rothbart, S.B.; Lu, R.; Xu, B.; Chen, W.-Y.; Tripathy, A.; Rockowitz, S.; Zheng, D.; Patel, D.J.; Allis, C.D.; et al. An H3K36 Methylation-Engaging Tudor Motif of Polycomb-like Proteins Mediates PRC2 Complex Targeting. Mol. Cell 2013, 49, 571–582.
- Schmitges, F.W.; Prusty, A.B.; Faty, M.; Stützer, A.; Lingaraju, G.M.; Aiwazian, J.; Sack, R.; Hess, D.; Li, L.; Zhou, S.; et al. Histone Methylation by PRC2 Is Inhibited by Active Chromatin Marks. Mol. Cell 2011, 42, 330–341.
- Yuan, W.; Xu, M.; Huang, C.; Liu, N.; Chen, S.; Zhu, B. H3K36 Methylation Antagonizes PRC2-Mediated H3K27 Methylation. J. Biol. Chem. 2011, 286, 7983–7989.
- Finogenova, K.; Bonnet, J.; Poepsel, S.; Schäfer, I.B.; Finkl, K.; Schmid, K.; Litz, C.; Strauss, M.; Benda, C.; Müller, J. Structural Basis for PRC2 Decoding of Active Histone Methylation Marks H3K36me2/3. eLife 2020, 9, e61964.
- Van Kruijsbergen, I.; Hontelez, S.; Veenstra, G.J.C. Recruiting Polycomb to Chromatin. Int. J. Biochem. Cell Biol. 2015, 67, 177–187.
- Ballaré, C.; Lange, M.; Lapinaite, A.; Martin, G.M.; Morey, L.; Pascual, G.; Liefke, R.; Simon, B.; Shi, Y.; Gozani, O.; et al. Phf19 Links Methylated Lys36 of Histone H3 to Regulation of Polycomb Activity. Nat. Struct. Mol. Biol. 2012, 19, 1257–1265.
- Blanco, E.; González-Ramírez, M.; Alcaine-Colet, A.; Aranda, S.; Croce, L.D. The Bivalent Genome: Characterization, Structure, and Regulation. Trends Genet. 2020, 36, 118–131.
- Zhang, Q.; Guan, P.; Zhao, L.; Ma, M.; Xie, L.; Li, Y.; Zheng, R.; Ouyang, W.; Wang, S.; Li, H.; et al. Asymmetric Epigenome Maps of Subgenomes Reveal Imbalanced Transcription and Distinct Evolutionary Trends in Brassica Napus. Mol. Plant 2021, 14, 604–619.
- Zhao, K.; Kong, D.; Jin, B.; Smolke, C.D.; Rhee, S.Y. A Novel Bivalent Chromatin Associates with Rapid Induction of Camalexin Biosynthesis Genes in Response to a Pathogen Signal in Arabidopsis. elife 2021, 10, e69508.
- Hansen, K.H.; Bracken, A.P.; Pasini, D.; Dietrich, N.; Gehani, S.S.; Monrad, A.; Rappsilber, J.; Lerdrup, M.; Helin, K. A Model for Transmission of the H3K27me3 Epigenetic Mark. Nat. Cell Biol. 2008, 10, 1291–1300.
- Margueron, R.; Justin, N.; Ohno, K.; Sharpe, M.L.; Son, J.; Iii, W.J.D.; Voigt, P.; Martin, S.; Taylor, W.R.; Marco, V.D.; et al. Role of the Polycomb Protein EED in the Propagation of Repressive Histone Marks. Nature 2009, 461, 762–767.
- Exner, V.; Aichinger, E.; Shu, H.; Wildhaber, T.; Alfarano, P.; Caflisch, A.; Gruissem, W.; Köhler, C.; Hennig, L. The Chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 Is Essential for H3K27me3 Binding and Function during Arabidopsis Development. PLoS ONE 2009, 4, e0005335.
- Cooper, S.; Dienstbier, M.; Hassan, R.; Schermelleh, L.; Sharif, J.; Blackledge, N.P.; De Marco, V.; Elderkin, S.; Koseki, H.; Klose, R.; et al. Targeting Polycomb to Pericentric Heterochromatin in Embryonic Stem Cells Reveals a Role for H2AK119u1 in PRC2 Recruitment. Cell Rep. 2014, 7, 1456–1470.
- Dobrinić, P.; Szczurek, A.T.; Klose, R.J. PRC1 Drives Polycomb-Mediated Gene Repression by Controlling Transcription Initiation and Burst Frequency. Nat. Struct. Mol. Biol. 2021, 28, 811–824.
- Blackledge, N.P.; Rose, N.R.; Klose, R.J. Targeting Polycomb Systems to Regulate Gene Expression: Modifications to a Complex Story. Nat. Rev. Mol. Cell Biol. 2015, 16, 643–649.
- Barbour, H.; Daou, S.; Hendzel, M.; Affar, E.B. Polycomb Group-Mediated Histone H2A Monoubiquitination in Epigenome Regulation and Nuclear Processes. Nat. Commun. 2020, 11, 5947.
- Yin, X.; Romero-Campero, F.J.; de Los Reyes, P.; Yan, P.; Yang, J.; Tian, G.; Yang, X.; Mo, X.; Zhao, S.; Calonje, M.; et al. H2AK121ub in Arabidopsis Associates with a Less Accessible Chromatin State at Transcriptional Regulation Hotspots. Nat. Commun. 2021, 12, 315.
More
