Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Kangkang Niu.
In eukaryotes, mRNAs translation is mainly mediated in a cap-dependent or cap-independent manner. The latter is primarily initiated at the internal ribosome entry site (IRES) in the 5′-UTR of mRNAs. It has been reported that the G-quadruplex structure (G4) in the IRES elements could regulate the IRES activity.
- G-quadruplex
- VEGFA
- IRES
- mRNA translation
1. Introduction
In protein synthesis, the genetic information in mRNA is translated into an amino acid sequence with the help of ribosomes and transfer RNAs (tRNAs) [1]. The mRNA translation is mediated in either a cap-dependent or cap-independent manner [2,3][2][3]. The former is a main regulation mode of mRNA translation in eukaryotes. However, when cells are suffering environmental stress, such as heat shock, hypoxia, amino acid deficiency, and viral infection, the cellular responses immediately lead to global repression of cap-dependent protein synthesis. Meanwhile, alternative mechanisms are activated to support the translation of specific mRNAs [4,5][4][5].
The mRNA translation can be subjectively separated into three stages: initiation, extension, and termination. The main difference between cap-dependent and cap-independent translation lies in the initiation stage. The initial step of cap-dependent translation initiation involves the binding of ribosomes, together with tRNAMet, to the 7-methylguanosine (m7-GpppG) 5′-cap of an mRNA. However, some viral and cellular mRNAs contain an internal ribosome entry site (IRES) that attracts ribosomes directly to the interior of the RNA to initiate cap-independent translation, which is also known as IRES mediated translation. IRES-mediated translation was first found in the RNAs of the virus [6,7][6][7]. Virus mRNA are uncapped and translated in a cap-independent manner. Eukaryotic cellular IRES elements were also identified, especially in the 5′-UTR of mRNA of some proto-oncogenes [8,9,10][8][9][10]. When global translation is blocked in response to stress to save energy, these genes were translated in the IRES dependent way to synthesize necessary proteins [3,11][3][11]. The mRNA IRES sequence can form a variety of RNA structures, such as stem loop, hairpin, G4 structures, and other secondary or tertiary structures. These advanced RNA structures can replace the 5′-cap of mRNA and recruit ribosomes to initiate the translation process [12,13][12][13]. The G4 structure is a special secondary structure of DNA and RNA. When nucleic acid sequences enrich in guanine (G), these sequences are favorable to form G planes, which consist of four Gs linked together through Hoogsteen hydrogen bond, and two or more G planes are stacked to form the G4 structure [14,15,16,17][14][15][16][17]. The G4 structure in the IRES sequence was found to directly bind with the 40S small subunit of ribosome, implying its regulatory role in the IRES-mediated translation [18].
The human vascular endothelial growth factor (VEGF) is a homodimeric glyco protein which plays critical roles in vasculogenesis and angiogenesis. The VEGF family consists VEGF-A (hereafter called VEGFA), VEGF-B, VEGF-C, VEGF-D, VEGF-E (viral VEGF), VEGF-F (snake venom VEGF), placenta growth factor (PlGF), and the endocrine gland-derived vascular endothelial growth factor (EG-VEGF). More attention was focused on the VEGFA due to its key roles in tumor growth and metastasis and eye disease [19]. The 5′UTR of VEGFA is G-rich and harbors two separate IRES sites (IRES-A at 749–1038 nt and IRES-B at 91–554 nt) that facilitate VEGFA translation initiation in a cap-independent manner to respond to various cellular stresses [20,21][20][21]. It has been reported that IRES-A could initiate VEGFA translation at AUG and synthesize a secreted form of VEGFA, which is crucial for the tumor angiogenesis [22]. Moreover, a G4 structure was identified in IRES-A region and the G4 structure is needed for VEGFA IRES mediated translation [23]. Consequently, the IRES-A of VEGFA was used to investigate the specific mechanism of G4 structures in regulating mRNA translation.
Human RNA-binding motif protein 4 (RBM4), a homologue of LARK in Bombyx mori, is ubiquitously expressed in human tissues [24]. WeAuthors previously demonstrated that LARK or RBM4 is a G4-binding protein in silkworms, mice, and human [25]. Studies have shown that RBM4 is mainly involved in biological processes, such as selective RNA splicing and RNA translation [26,27][26][27]. RBM4 has been reported to inhibit cap-dependent translation but promote the IRES-mediated translation under cellular stress [24,26][24][26]. However, how RBM4, as a G4-binding protein, participates in IRES-mediated translation remains to be elucidated.
2. IRES-A Promoted the VEGFA mRNA Translation in 293T Cells
To investigate whether the IRES-A element of VEGFA 5′-UTR mediates mRNA translation, the dual luciferase reporter psiCHECK2 was applied to detect its function in mRNA translation. The HSV-TK promoter upstream of the firefly luciferase (FLuc) marker gene was replaced by the VEGFA IRES-A sequence to generate a pR-IRES-F vector. The control pRF was the empty vector without the HSV-TK promoter (Figure 1A). The pR-IRES-F plasmid was used to transfect the 293T cells, followed by a luciferase activity assay after 24 h. The results showed that VEGFA IRES-A activity (FLuc/RLuc) was significantly increased, as compared to the control plasmid pRF (Figure 1B). Further analysis showed that the RLuc activity was not significantly changed, while FLuc activity was significantly increased (Figure 1C). These results suggest that the VEGFA IRES-A could promote either the transcription or translation of FLuc in the 293T cells.
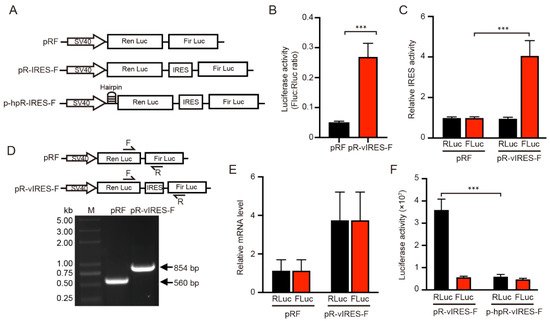

Figure 1. Dual luciferase assay for the functional analysis of VEGFA IRES-A (vIRES). (A) Schematic illustration of the dicistronic reporter gene vector constructs used in this study. (B,C) Influence of vIRES positioned upstream the firefly and renilla luciferases. The IRES activity are represented as ratios of firefly to renilla luciferase. The individual luciferase activity was normalized to that of the control vector pRF. (D) RT-PCR and (E) qRT-PCR verification of the influence of vIRES on the mRNA splicing and transcription. (F) Activity of the individual luciferases in the bicistronic vectors in the absence or presence of a hairpin upstream of renilla luciferase. *** indicates significant difference at p < 0.001.
Various mechanisms could account for the increase of Fluc translation downstream of the dicistronic mRNA. The dicistronic mRNA may be spliced into monocistronic mRNAs. The inserted IRES sequence may contain an element that promotes the read-through past the RLuc cistron and causes reinitiation of Fluc cistron. Finally, the inserted sequence may act as an IRES site mediating cap-independent translation. In order to determine whether the effect of the VEGFA IRES-A on the increase of the FLuc activity occurs at the mRNA translation level or transcriptional level, and to further verify that RLuc and FLuc are translated from the same mRNA, the upstream primer and downstream primer within the RLuc and FLuc genes respectively were specifically designed to detect whether the mRNA was spliced and the mRNA level of individual genes was examined. Total RNA were extracted from the 293T cells transfected with pRF and PR-IRES-F plasmids and reverse transcribed into cDNA. PCR was then performed to detect whether mRNA is spliced between the RLuc and FLuc mRNA. The results showed that the resultant PCR band was a single one, the size was expected, and there was no cleavage between RLuc and FLuc mRNAs in both the pRF and PR-IRES-F cases (Figure 1D). In addition, the qPCR results showed that there was no difference in mRNA expression between RLuc and FLuc in both the pRF and PR-IRES-F vectors (Figure 1E). These results indicated that the protein expression (as indicated by luciferase activity) of RLuc and FLuc was the result of the translation of the same mRNA without splicing.
Besides, to verify that the FLuc translation was not caused by mRNA read-through during RLuc translation, weauthors added a hairpin structure upstream of the RLuc sequence (Figure 1A). This hairpin structure significantly inhibited the RLuc activity, whereas the FLuc activity was not impacted (Figure 1F). If the insertion could enhance ribosomal read-through, the Fluc activity should be reduced by an equivalent amount. Thus, given that the promoter (SV40) of the two vector constructs was the same, the significant increase in the FLuc enzyme activity was considered to be the result of the increased IRES mediated translation.
3. IRES Activity Was Affected by G4 Structure
It has been demonstrated by DNAase I footprinting that the 774~90 nt region of the IRES-A sequence in the VEGFA 5′-UTR could form a G4 structure and this G4 structure had a regulatory effect on the IRES activity [23]. In this study, the CD experiments revealed that the oligonucleotide of this mRNA region had a maximum absorption peak at 264 nm, and the peak was increased when 100 mM K+ was added (Figure 2A), presenting a classical peak type of parallel G4 structure [28,29][28][29]. When the G4 sequence was mutated, the maximum absorption peak shifted to 275 nm, and the K+-dependent increase of the absorption peak disappeared (Figure 2A), indicating that the mutant RNA could not form G4 structures any more consistent than previously reported [28,29][28][29]. The G4 mutation resulted in a significant decrease in the FLuc activity, but not in the RLuc activity (Figure 2B). Moreover, when the cells were treated with 2.5 μM PDS, a small molecular compound known to bind and stabilize the G4 structure, the FLuc activity was significantly increased, as compared to the control (DMSO), while the Rluc activity was not significantly changed (Figure 2C). When the G4 sequence was mutated, the FLuc activity did not respond to the PDS treatment (Figure 2D). These results suggested that the IRES-A element could form a G4 structure that facilitated the IRES-mediated FLuc mRNA translation.
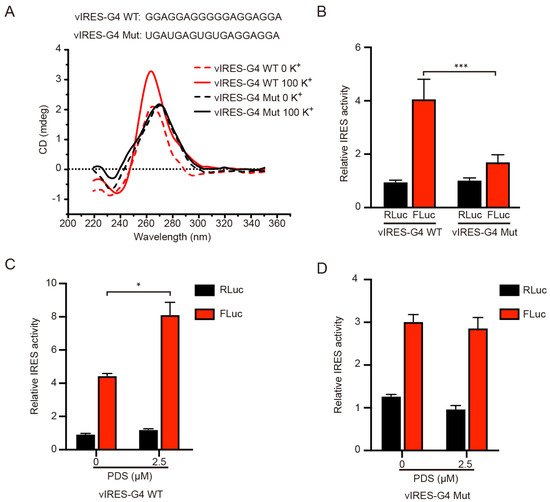

Figure 2. Effect of G4 structure on the individual luciferase activity. (A) CD analysis of the wild type and mutant G4 sequence in the vIRES vector in the absence or presence of 100 mM K+. (B) The influence of the G4 mutation on the FLuc luciferase activity. PDS treatment significantly enhanced the FLuc luciferase activity of the wild type G4 (C), but not the mutant G4 (D). * indicates significant difference at p < 0.05; *** indicates significant difference at p < 0.001.
4. RBM4 Specifically Bound the G4 Structure in IRES Element
To verify whether RBM4 binds the G4 structure in the VEGFA IRES-A, RNA G4 oligonucleotides were synthesized in vitro and annealed to allow the formation of the G4 structure, followed by EMSA with purified RBM4 protein. The result showed that RBM4 could specifically bind the G4 structure (Figure 3A, lane 2) and this specific binding could be competed off gradually by the unlabeled probe (Figure 3A, lane 3 and 4). It is noted that there were four binding bands, implying that various G4 structure isforms or formats of the interaction between the G4 structure and the protein might be present in the binding system. The mutated RNA G4 probe could not bind the RBM4 protein (Figure 3A, lane 5). When the protein content of RBM4 was increased gradually, the binding intensity was strengthened (Figure 3B). These results indicated that RBM4 could specifically bind the G4 structure in the VEGFA IRES-A element.
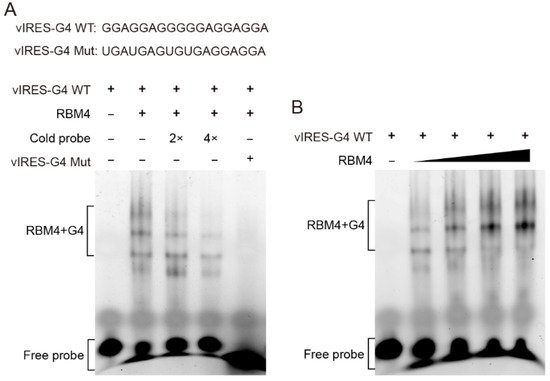

Figure 3. RBM4 specifically bound to the G4 structure in vIRES. (A) EMSA analysis of the binding between RBM4 and the G4 structure in the vIRES. (B) The binding between RBM4 and G4 was gradually enhanced with the increase of RBM4 protein level.
5. RBM4 Promoted the VEGFA IRES-A Activity
To investigate the effect of RBM4 on the IRES-A translation activity, we conducted RBM4 RNAi and over-expression in the 293T cells. The FLuc enzyme activity was significantly decreased when the RBM4 was down-regulated, as compared to that of the control (Figure 4A,B). The RBM4 RNAi did not affect the FLuc activity when the G4 sequence was mutated (Figure 4B). Contrarily, the FLuc activity was notably increased when RBM4 protein was over-expressed (Figure 4C,D). Similarly, the FLuc enzyme activity did not change when the G4 sequence was mutated (Figure 4D). Taken together, these results suggest that RBM4 could specifically bind the G4 structure in the VEGFA IRES-A and enhance the IRES-mediated translation of VEGFA mRNA.
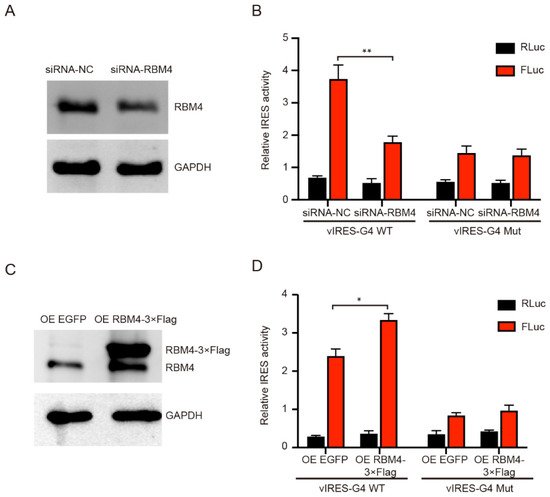

Figure 4. Effect of RBM4 on the expression of luciferases. (A) Western blot analysis of RBM4 protein after RNAi. (B) Knockdown of RBM4 inhibited the FLuc luciferase activity. (C) Western blot analysis of RBM4 protein after over-expression. (D) Over-expression of RBM4 resulted in the increase in the FLuc luciferase activity in the wild type IRES-A element, but not in the mutated element. * indicates significant difference at p < 0.05; ** indicates significant difference at p < 0.01.
References
- Gebauer, F.; Hentze, M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004, 5, 827–835.
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745.
- Yang, Y.; Wang, Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 2019, 11, 911–919.
- Han, S.; Sun, S.; Li, P.; Liu, Q.; Zhang, Z.; Dong, H.; Sun, M.; Wu, W.; Wang, X.; Guo, H. Ribosomal Protein L13 Promotes IRES-Driven Translation of Foot-and-Mouth Disease Virus in a Helicase DDX3-Dependent Manner. J. Virol. 2020, 94, e01679-19.
- Kampen, K.R.; Sulima, S.O.; Verbelen, B.; Girardi, T.; Vereecke, S.; Rinaldi, G.; Verbeeck, J.; de Beeck, J.O.; Uyttebroeck, A.; Meijerink, J.P.P.; et al. The ribosomal RPL10 R98S mutation drives IRES-dependent BCL-2 translation in T-ALL. Leukemia 2019, 33, 319–332.
- Jang, S.K.; Krausslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988, 62, 2636–2643.
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325.
- Stoneley, M.; Paulin, F.E.; Le Quesne, J.P.; Chappell, S.A.; Willis, A.E. C-Myc 5’ untranslated region contains an internal ribosome entry segment. Oncogene 1998, 16, 423–428.
- Coldwell, M.J.; Mitchell, S.A.; Stoneley, M.; MacFarlane, M.; E Willis, A. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 2000, 19, 899–905.
- Leppek, K.; Das, R.; Barna, M. Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174.
- Lampe, S.; Kunze, M.; Scholz, A.; Brauß, T.F.; Winslow, S.; Simm, S.; Keller, M.; Heidler, J.; Wittig, I.; Brüne, B.; et al. Identification of the TXNIP IRES and characterization of the impact of regulatory IRES trans-acting factors. Biochim. Biophys. Acta. Gene Regul. Mech. 2018, 1861, 147–157.
- Lozano, G.; Martínez-Salas, E. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015, 12, 113–120.
- Jodoin, R.; Carrier, J.C.; Rivard, N.; Bisaillon, M.; Perreault, J.-P. G-quadruplex located in the 5’UTR of the BAG-1 mRNA affects both its cap-dependent and cap-independent translation through global secondary structure maintenance. Nucleic Acids Res. 2019, 47, 10247–10266.
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA 1962, 48, 2013–2018.
- Yaku, H.; Fujimoto, T.; Murashima, T.; Miyoshi, D.; Sugimoto, N. Phthalocyanines: A new class of G-quadruplex-ligands with many potential applications. Chem. Commun. Camb. 2012, 48, 6203–6216.
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637.
- Bartas, M.; Brázda, V.; Karlický, V.; Červeň, J.; Pečinka, P. Bioinformatics analyses and in vitro evidence for five and six stacked G-quadruplex forming sequences. Biochimie 2018, 150, 70–75.
- Bhattacharyya, D.; Diamond, P.; Basu, S. An Independently folding RNA G-quadruplex domain directly recruits the 40S ribosomal subunit. Biochemistry 2015, 54, 1879–1885.
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264.
- Huez, I.; Créancier, L.; Audigier, S.; Gensac, M.C.; Prats, A.C.; Prats, H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell Biol. 1998, 18, 6178–6190.
- Miller, D.L.; Dibbens, J.A.; Damert, A.; Risau, W.; Vadas, M.A.; Goodall, G.J. The vascular endothelial growth factor mRNA contains an internal ribosome entry site. FEBS Lett. 1998, 434, 417–420.
- Bornes, S.; Boulard, M.; Hieblot, C.; Zanibellato, C.; Iacovoni, J.S.; Prats, H.; Touriol, C. Control of the vascular endothelial growth factor internal ribosome entry site (IRES) activity and translation initiation by alternatively spliced coding sequences. J. Biol. Chem. 2004, 279, 18717–18726.
- Morris, M.J.; Negishi, Y.; Pazsint, C.; Schonhoft, J.D.; Basu, S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010, 132, 17831–17839.
- Markus, M.A.; Morris, B.J. RBM4: A multifunctional RNA-binding protein. Int. J. Biochem. Cell Biol. 2009, 41, 740–743.
- Niu, K.K.; Xiang, L.J.; Jin, Y.; Peng, Y.; Wu, F.; Tang, W.; Zhang, X.; Deng, H.; Xiang, H.; Li, S.; et al. Identification of LARK as a novel and conserved G-quadruplex binding protein in invertebrates and vertebrates. Nucleic Acids Res. 2019, 47, 7306–7320.
- Lin, J.C.; Hsu, M.; Tarn, W.Y. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc. Natl. Acad. Sci. USA 2007, 104, 2235–2240.
- Wang, Y.; Chen, D.; Qian, H.; Tsai, Y.S.; Shao, S.; Liu, Q.; Dominguez, D.; Wang, Z. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell 2014, 26, 374–389.
- Shahid, R.; Bugaut, A.; Balasubramanian, S. The BCL-2 5′ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry 2010, 49, 8300–8306.
- Huang, H.; Zhang, J.; Harvey, S.E.; Hu, X.; Cheng, C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017, 31, 2296–2309.
More
