Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Beatrix Zheng and Version 2 by Beatrix Zheng.
Pirin, an evolutionary conserved non-heme Fe-containing family member of the cupin superfamily of proteins, is regulated by Nrf2 in human and mouse cells and in the mouse colon in vivo. Moreover, pirin is overexpressed in human colorectal tumours where pirin expression correlates with Nrf2 activation, suggesting Nrf2 dependence. The depletion of pirin in the human colorectal cancer cell line DLD1 does not affect cell viability or migration. Understanding the functional consequences of the observed pirin upregulation in colorectal cancer requires further investigation.
- AKR1B10
- AKR1C1
- colorectal cancer
- DLD1
- Nrf2
- NQO1
- pirin
1. Introduction
Pirin is a highly evolutionary conserved non-heme Fe-containing family member of the cupin superfamily of proteins, which in its oxidized (Fe+++-bound) form, binds to the RelA (p65) subunit and enhances the activity of transcription factor NFκB [1]. Pirin can also form complexes with B-cell lymphoma 3-encoded protein (Bcl3) and NFκB1 (p50) on NFκB DNA binding sites [2]. Furthermore, several reports have implicated pirin as an important factor in cancer cell proliferation, migration, and tumour progression in the context of melanoma, breast, lung, cervical, prostate, and oral cancer [3][4][5][6][7].Although the molecular details of how pirin is regulated are incompletely understood, increasing evidence points to the role of transcription factor nuclear factor erythroid 2 p45-related factor 2 (Nrf2, gene name NFE2L2) in mediating the gene expression of pirin. Thus, a gene expression study comparing the small airway epithelium of healthy non-smokers to that of healthy smokers using samples obtained by fiberoptic bronchoscopy identified pirin as a “smoking-responsive Nrf2-modulated gene”, and further showed that the promoter region of the human PIR gene contains two antioxidant response elements (AREs), the DNA sequences to which Nrf2 binds, located at −3209 bp and −5566 bp upstream of the transcription start site [8]. It was also reported that the basal expression of pirin in HeLa cells is regulated by Nrf2 [9]. A chemical proteomics approach suggested pirin as one of the proteins regulated by Nrf2 in non-small cell lung cancer cell lines [10]. Recent studies have implicated Nrf2 in the overexpression of pirin caused by the high-risk human papillomavirus (HR-HPV) E7 oncoprotein [6], as well as by small-molecule inducers of ferroptosis [11]. Interestingly, RNA-sequencing (RNA-seq) expression profiling of HEK293T cells has shown that pirin expression is induced upon treatment with the electrophile 4-hydroxynonenal (HNE), but not by the oxidant H2O2 [12]. Both HNE and H2O2 induce Nrf2-mediated transcription following chemical modification of specific cysteine sensors of Kelch-like ECH associated protein 1 (Keap1), the principal negative regulator of Nrf2 [13][14]. Curiously, inhibition of the expression of pirin was seen when cervical carcinoma cells were exposed to curcumin [15], a dietary plant Michael reaction acceptor, which we and others have found to activate Nrf2 [16][17][18].
2. Pirin Is a Transcriptional Target of Nrf2 in Human Cell Lines
The research first compared the mRNA levels for pirin in the human colorectal cancer cell line DLD1 (Nrf2-WT) as well as Nrf2-knockout (Nrf2-KO) and Nrf2-gain-of-function (Nrf2-GoF) mutant isogenic lines that had been generated using CRISPR/Cas9 genome editing [19]. In comparison with Nrf2-WT, the mRNA levels for pirin were 2-fold higher in Nrf2-GoF DLD1 cells, and 50% lower in their Nrf2-knockout counterparts (Figure 1A). This pattern of expression of pirin was similar to that of the classical Nrf2 target protein NAD(P)H:quinone oxidoreductase 1 (NQO1) (Figure 1B), confirming Nrf2 activation. Similar to NQO1, the mRNA levels for pirin were increased 1.5-fold by exposure to TBE-31, and this increase was diminished in Nrf2-knockout DLD1 cells (Figure 1A). The protein levels of pirin were higher in Nrf2-gain-of-function DLD1 cells than in their WT counterparts, and treatment with both TBE-31 and the classical Nrf2 inducer sulforaphane increased the protein levels of pirin in WT cells with no further effect in Nrf2-gain-of-function cells (Figure 1C).
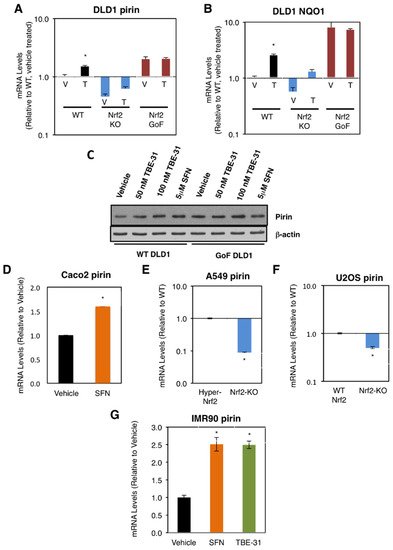

Figure 1. Pirin is a transcriptional target of Nrf2 in human cells. (A,B) mRNA levels for pirin (A) and NQO1 (B) in Nrf2-wild-type (WT), Nrf2-knockout (Nrf2-KO), and Nrf2 gain-of-function mutant (Nrf2-GoF) human DLD1 colorectal cancer cells treated with vehicle (V) or 10 nM TBE-31 (T) for 16 h. (C) Protein levels for pirin in Nrf2-wild-type (WT) and Nrf2 gain-of-function mutant (Nrf2-GoF) human DLD1 colorectal cancer cells treated with vehicle, 50 nM TBE-31, 100 nM TBE-31, or 5 μM sulforaphane (SFN) for 16 h. (D) mRNA levels for pirin in Caco2 colorectal cancer cells treated with vehicle or 5 μM SFN for 24 h. (E) mRNA levels for pirin in A549 cells (high Nrf2 levels, “Hyper-Nrf2”) and their Nrf2-knockout (Nrf2-KO) mutant counterparts. (F) mRNA levels for pirin in U2OS cells (normal Nrf2 levels, WT Nrf2) and their Nrf2-knockout (Nrf2-KO) mutant counterparts. (G) mRNA levels for pirin in IMR90 normal human lung fibroblasts treated with vehicle, 5 μM SFN or 100 nM TBE-31 for 16 h. * p < 0.05. The TaqMan data were normalized using human hypoxanthine phosphoribosyltransferase 1 (Hprt1) as an internal control.
Sulforaphane treatment also increased (by 1.6-fold) the mRNA levels for pirin in the colorectal cancer cell line Caco2 (Figure 1D). Conversely, the mRNA levels for pirin decreased by 90% upon knockout of Nrf2 in the human lung cancer cell line A549, which has constitutively high levels of Nrf2 due to a mutation in Keap1 and hypermethylation of its promoter (Figure 1E). Similarly, the mRNA levels for pirin decreased by 50% upon knockout of Nrf2 in the human osteosarcoma cell line U2OS, in which the levels of Keap1 and Nrf2 are normal (Figure 1F).
In addition to cancer cells, the dependence on Nrf2 for the expression of pirin was also observed in normal human lung fibroblast IMR90 cells, where treatment with TBE-31 or sulforaphane caused similar levels of induction (by ~2.5-fold) of the expression of pirin (Figure 1G). Collectively, these experiments establish the dependence of pirin expression on Nrf2 in human cells.
3. Pirin Is a Transcriptional Target of Nrf2 in Mice
The dependence on Nrf2 of both basal and inducible expression of pirin and NQO1 was also observed in primary cultures of mouse embryonic fibroblast (MEF) cells, intestinal organoids, and in the murine colon in vivo. For these experiments, we used wild-type (WT), Nrf2-knockout (Nrf2-KO, Nrf2−/−) and Keap1-knockdown (Keap1-KD, Keap1flox/flox) mice [20][21][22]. Compared to WT, the mRNA levels for pirin were 60% lower and not inducible by TBE-31 in Nrf2-knockout MEF cells, whereas these levels were 1.5-fold higher in their Keap1-knockdown counterparts (Figure 2A). The basal and TBE-31 induced expression of NQO1 in the three genotypes paralleled that of pirin (Figure 2B).
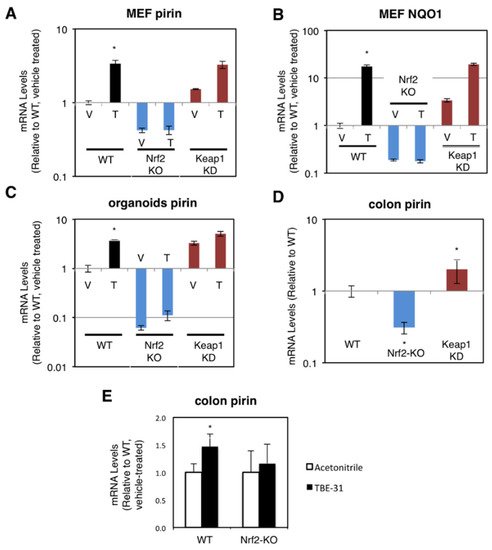

Figure 2. Pirin is a transcriptional target of Nrf2 in mouse cells and in vivo. (A,B) mRNA levels for pirin (A) and NQO1 (B) in Nrf2-wild-type (WT), Nrf2-knockout (Nrf2-KO), and Keap1-knockdown (Keap1-KD) mouse embryonic fibroblast (MEF) cells treated with vehicle (V) or 50 nM TBE-31 (T) for 24 h. The TaqMan data were normalized using eukaryotic 18S rRNA (18S) as an internal control. (C) mRNA levels for pirin in Nrf2-wild-type (WT), Nrf2-knockout (Nrf2-KO), and Keap1-knockdown (Keap1-KD) mouse intestinal organoid cultures treated with vehicle (V) or 10 nM TBE-31 (T) for 24 h. (D) mRNA levels for pirin in the colon of wild-type (WT), Nrf2-knockout (Nrf2-KO), and Keap1-knockdown (Keap1-KD) C57BL6 mice (n = 6–8). (E) mRNA levels for pirin in the colons of wild-type (WT) and Nrf2-knockout (Nrf2-KO) mice (n = 4–5), which had been treated with TBE-31 (5 nmol/g body weight, 3 times, at 24-h intervals, per os, black bars) or vehicle (0.7% DMSO in corn oil, white bars). * p < 0.05. The TaqMan data were normalized using mouse ribosomal protein lateral stalk subunit P0 (rplp0) as an internal control.
Next, organoids from the small intestine of WT, Nrf2-KO, and Keap1-KD mice were prepared. In agreement with the data in human and MEF cells, compared to WT, the mRNA levels for pirin were 3.3-fold higher in Keap1-knockdown organoids, whereas these levels were >90% lower in their Nrf2-knockout counterparts (Figure 2C). Treatment with TBE-31 induced pirin 3.9-fold in WT organoids, but this induction was greatly diminished in organoids from Nrf2-knockout or Keap1-knockdown mice (Figure 2C).
In the colons of mice from the three genotypes, the expression of pirin was 70% lower and 2-fold higher in Nrf2-knckout and Keap1-knckdown, respectively (Figure 2D). Oral administration of TBE-31 to mice induced the expression of pirin in wild-type animals, but had no effect in their Nrf2-knockout counterparts (Figure 2E). Thus, using genetic and pharmacologic approaches, we have demonstrated that pirin is a transcriptional target of Nrf2 in human colorectal cancer cells, mouse intestinal organoid cultures and in vivo in the murine colon.
4. Pirin Is Overexpressed in Human Colorectal Tumors
The authors determined the gene expression of pirin in 48 primary human colorectal tumours and matched normal tissue from the same patients. Using 18S as a reference gene and a 2-fold increase or a 50% decrease relative to normal tissue as a cut-off, we found that the mRNA levels for pirin were increased in 30/48 and decreased in 2/48 tumour samples in comparison with their corresponding matched normal tissues (Figure 3A). The mRNA levels for NQO1 were increased in 25/48 tumours and decreased in the same two tumour samples, which also had lower pirin expression (Figure 3B). Based on Spearman and Kendall rank correlation test, the folds change of pirin expression in tumour vs. the corresponding normal tissues correlated with the fold change of NQO1 expression (p = 0.001271 and p = 0.0008092). Furthermore, when we examined the mRNA levels for Nrf2 in the human tissues, we found them to be increased in 18/48 and decreased in 2/48 tumour samples in comparison with their matched normal tissues (Figure 3C). The correlation test also found that the fold change of pirin expression correlated with the fold change of Nrf2 expression (p = 1.49E-05 and p = 9.32E-06). Together, these data strongly suggest that the overexpression of pirin in the tumour tissues is dependent on Nrf2.
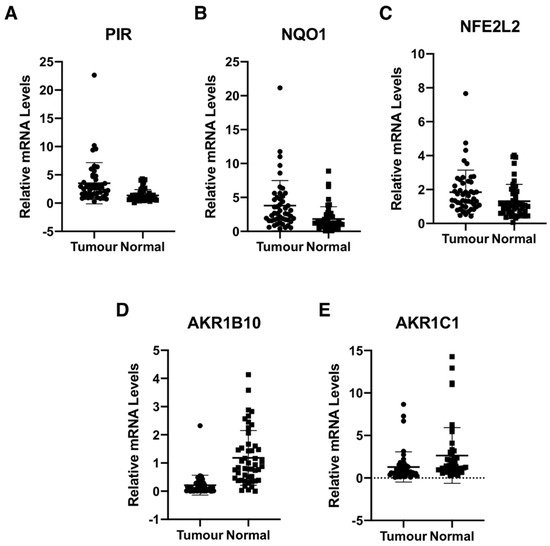

Figure 3. Pirin is overexpressed in human colorectal tumours. mRNA levels for pirin (A), NQO1 (B), NFE2L2 (C), AKR1B10 (D), and AKR1C1 (E) in matched tumour and normal tissues from human colorectal cancer patients (n = 48). p < 0.05 in all cases. The TaqMan data were normalized using eukaryotic 18S rRNA (18S) as an internal control.
Although Nrf2 is primarily regulated on the protein level, oncogene-induced Nrf2 overexpression has also been reported [23]. Thus, it has been shown that the expression of an endogenous oncogenic allele of K-Ras(G12D) in mouse embryonic fibroblasts increases the transcription of Nrf2 [23]. Therefore, we compared the expression of Nrf2, pirin, and NQO1 in K-RasWT/− and K-RasG13D/− DLD1 cell lines. We found that the mRNA levels for Nrf2 were 1.3-fold higher in the oncogenic K-Ras-expressing cell line compared to its wild-type K-Ras-expressing counterpart (Figure 4A), confirming the transcriptional upregulation of Nrf2 under conditions of K-Ras activation. The mRNA levels for pirin (Figure 4B) and NQO1 (Figure 4C) were also increased in the oncogenic K-Ras-expressing cells by 1.3- and 2.5-fold, respectively, in agreement with Nrf2 activation. Together with the analysis of the human tissues, these data indicate that Nrf2 is activated in human colorectal cancers, and this activation is likely mediated by both transcriptional and post-transcriptional mechanisms.
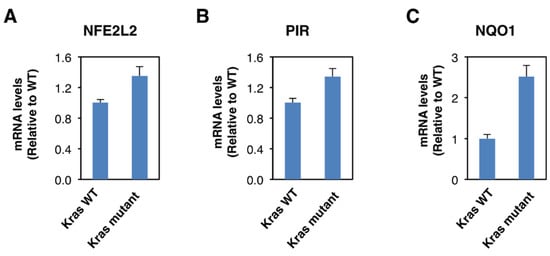

Figure 4. Constitutive activation of K-Ras enhances NFE2L2 transcription. mRNA levels for NFE2L2 (A), pirin (B), and NQO1 (C) in K-RasWT/− and K-RasG13D/− mutant DLD1 cell lines. p < 0.05 in all cases. The TaqMan data were normalized using eukaryotic 18S rRNA (18S) as an internal control.
Curiously however, the expression of the aldo-keto reductase family 1 members AKR1B10 and AKR1C1, which are well-established Nrf2 targets in humans, showed the opposite pattern to that of pirin, Nrf2 and NQO1 in the human tissues. Thus, compared to matched normal tissue, the mRNA levels for AKR1B10 and AKR1C1 were decreased in 39/48 and 28/48 tumour samples, respectively (Figure 3D,E). In both cases, the differences between the tumour and normal tissues were statistically significant. This finding suggests that transcriptional mechanism(s) other than regulation by Nrf2 are responsible for the downregulation of the gene expression AKR1B10 and AKR1C1 in human colorectal tumours.
5. Pirin Does Not Affect the Viability or Migration of DLD1 Colorectal Cancer Cells
The authors then addressed the potential functional consequences of the observed pirin upregulation in colorectal tumours. Several reports have implicated pirin as an important factor in cancer cell proliferation, migration, and tumour progression [3][4][5]. Thus, first we asked whether pirin affects the viability of DLD1 colorectal cancer cells using two approaches: (i) pharmacological, by treatment with triphenyl compound A (TPhA), a small molecule inhibitor of pirin [5], and (ii) genetic, by knockdown of pirin using siRNA. Exposure to TPhA concentration-dependently inhibited the viability of DLD1 cells (Figure 5A). By contrast, depleting pirin by siRNA had no effect on cell viability (Figure 5B), in spite of the high efficiency of the knockdown at both the mRNA (Figure 5C) and the protein (Figure 5D) levels. Furthermore, the TPhA-mediated inhibition of cell viability occurred at a similar level in cells transfected with pirin siRNA as it did in siRNA negative control-transfected cells (Figure 5E), strongly suggesting that the inhibitory effect of this compound in our experimental system was pirin-independent.
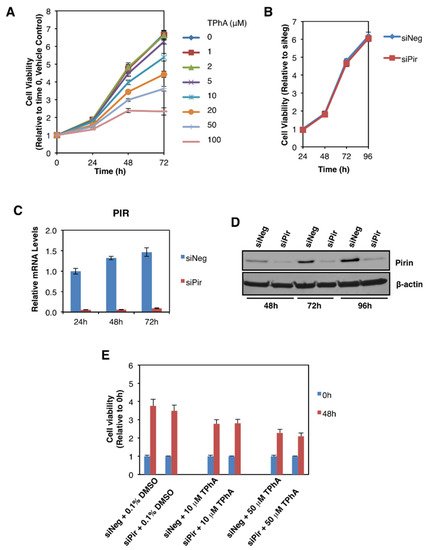

Figure 5. Pirin does not affect the viability of DLD1 colorectal cancer cells. (A) DLD1 cells (5 × 103 per well) plated in 96-well plates were treated with the indicated concentration of TphA for 24-, 48- and 72 h. Cell viability was assessed using the Alamar Blue fluorometric assay. (B) DLD1 cells (5 × 103 per well) were transfected with 20 nM ON-TARGET plus Smart Pool siRNA against human PIR (siPir) or ON-TARGET plus Non-targeting Control Pool (siNeg). Cell viability was monitored for four days after transfection with Alamar Blue reagent. (C) mRNA levels for pirin in DLD1 cells transfected with 20 nM siNeg or siPir for 24-, 48- and 72 h. (D) Protein levels for pirin in DLD1 cells transfected with 20 nM siNeg or siPir for 48-, 72- and 96 h. (E) DLD1 cells (5 × 103 per well) were transfected with 20 nM siNeg or siPir for 48 h, and then treated with 0.1% DMSO, 10 μM or 50 μM TPhA. Cell viability was assessed using the Alamar Blue fluorometric assay at 0- and 48 h after treatment.
The authors then asked whether pirin affects the migration of DLD1 colorectal cancer cells using a well-established functional wound-healing assay. To this end, cells were transfected with pirin siRNA or siRNA negative control as above, grown to confluency, and subsequently serum starved to prevent cell proliferation. A uniform scratch wound was then introduced in the confluent cell monolayer in a tightly-controlled manner, and the resulting wound closure was monitored by real-time imaging. Depleting pirin by siRNA had no significant effect on wound closure (Figure 6), suggesting that under these experimental conditions, pirin does not affect cell migration.
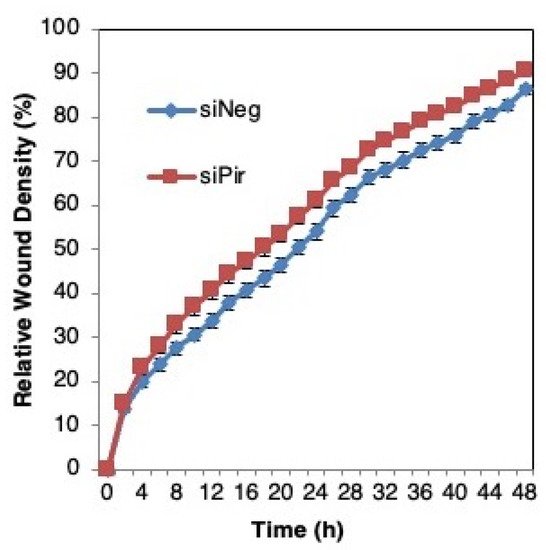

Figure 6. Pirin does not affect the migration of DLD1 colorectal cancer cells. DLD1 cells were transfected with 20 nM ON-TARGET plus Smart Pool siRNA against human PIR (siPir) or ON-TARGET plus Non-targeting Control Pool (siNeg) and grown for 48 h. Transfected cells (1.5 × 104 cells per well, 6 replicate wells for each condition) were seeded in 96-well ImageLock plates (Essen BioScience) and grown for a further 48 h to confluence. Cell migration was monitored at 2-h intervals for two days using IncuCyte™.
References
- Liu, F.; Rehmani, I.; Esaki, S.; Fu, R.; Chen, L.; de Serrano, V.; Liu, A. Pirin is an iron-dependent redox regulator of NF-kappaB. Proc. Natl. Acad. Sci. USA 2013, 110, 9722–9727.
- Dechend, R.; Hirano, F.; Lehmann, K.; Heissmeyer, V.; Ansieau, S.; Wulczyn, F.G.; Scheidereit, C.; Leutz, A. The Bcl-3 oncoprotein acts as a bridging factor between NF-kappaB/Rel and nuclear co-regulators. Oncogene 1999, 18, 3316–3323.
- Suleman, M.; Chen, A.; Ma, H.; Wen, S.; Zhao, W.; Lin, D.; Wu, G.; Li, Q. PIR promotes tumorigenesis of breast cancer by upregulating cell cycle activator E2F1. Cell Cycle 2019, 18, 2914–2927.
- Licciulli, S.; Luise, C.; Zanardi, A.; Giorgetti, L.; Viale, G.; Lanfrancone, L.; Carbone, R.; Alcalay, M. Pirin delocalization in melanoma progression identified by high content immuno-detection based approaches. BMC Cell Biol. 2010, 11, 5.
- Miyazaki, I.; Simizu, S.; Okumura, H.; Takagi, S.; Osada, H. A small-molecule inhibitor shows that pirin regulates migration of melanoma cells. Nat. Chem. Biol. 2010, 6, 667–673.
- Carrillo-Beltran, D.; Munoz, J.P.; Guerrero-Vasquez, N.; Blanco, R.; Leon, O.; Lino, V.D.; Tapia, J.C.; Maldonado, E.; Dubois-Camacho, K.; Hermoso, M.A.; et al. Human Papillomavirus 16 E7 Promotes EGFR/PI3K/AKT1/NRF2 Signaling Pathway Contributing to PIR/NF-kappa B Activation in Oral Cancer Cells. Cancers 2020, 12, 1904.
- Arai, T.; Kojima, S.; Yamada, Y.; Sugawara, S.; Kato, M.; Yamazaki, K.; Naya, Y.; Ichikawa, T.; Seki, N. Pirin: A potential novel therapeutic target for castration-resistant prostate cancer regulated by miR-455-5p. Mol. Oncol. 2019, 13, 322–337.
- Hubner, R.H.; Schwartz, J.D.; De Bishnu, P.; Ferris, B.; Omberg, L.; Mezey, J.G.; Hackett, N.R.; Crystal, R.G. Coordinate control of expression of Nrf2-modulated genes in the human small airway epithelium is highly responsive to cigarette smoking. Mol. Med. 2009, 15, 203–219.
- Brzoska, K.; Stepkowski, T.M.; Kruszewski, M. Basal PIR expression in HeLa cells is driven by NRF2 via evolutionary conserved antioxidant response element. Mol. Cell. Biochem. 2014, 389, 99–111.
- Bar-Peled, L.; Kemper, E.K.; Suciu, R.M.; Vinogradova, E.V.; Backus, K.M.; Horning, B.D.; Paul, T.A.; Ichu, T.A.; Svensson, R.U.; Olucha, J.; et al. Chemical Proteomics Identifies Druggable Vulnerabilities in a Genetically Defined Cancer. Cell 2017, 171, 696–709.e23.
- Hu, N.; Bai, L.; Dai, E.; Han, L.; Kang, R.; Li, H.; Tang, D. Pirin is a nuclear redox-sensitive modulator of autophagy-dependent ferroptosis. Biochem. Biophys. Res. Commun. 2021, 536, 100–106.
- Poganik, J.R.; Long, M.J.C.; Disare, M.T.; Liu, X.; Chang, S.H.; Hla, T.; Aye, Y. Post-transcriptional regulation of Nrf2-mRNA by the mRNA-binding proteins HuR and AUF1. FASEB J. 2019, 33, 14636–14652.
- McMahon, M.; Lamont, D.J.; Beattie, K.A.; Hayes, J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA 2010, 107, 18838–18843.
- Suzuki, T.; Muramatsu, A.; Saito, R.; Iso, T.; Shibata, T.; Kuwata, K.; Kawaguchi, S.I.; Iwawaki, T.; Adachi, S.; Suda, H.; et al. Molecular Mechanism of Cellular Oxidative Stress Sensing by Keap1. Cell Rep. 2019, 28, 746–758.e4.
- Aedo-Aguilera, V.; Carrillo-Beltran, D.; Calaf, G.M.; Munoz, J.P.; Guerrero, N.; Osorio, J.C.; Tapia, J.C.; Leon, O.; Contreras, H.R.; Aguayo, F. Curcumin decreases epithelialmesenchymal transition by a Pirindependent mechanism in cervical cancer cells. Oncol. Rep. 2019, 42, 2139–2148.
- Dinkova-Kostova, A.T.; Talalay, P. Relation of structure of curcumin analogs to their potencies as inducers of Phase 2 detoxification enzymes. Carcinogenesis 1999, 20, 911–914.
- Ashrafizadeh, M.; Ahmadi, Z.; Mohammadinejad, R.; Farkhondeh, T.; Samarghandian, S. Curcumin Activates the Nrf2 Pathway and Induces Cellular Protection Against Oxidative Injury. Curr. Mol. Med. 2020, 20, 116–133.
- Park, J.Y.; Sohn, H.Y.; Koh, Y.H.; Jo, C. Curcumin activates Nrf2 through PKCdelta-mediated p62 phosphorylation at Ser351. Sci. Rep. 2021, 11, 8430.
- Torrente, L.; Sanchez, C.; Moreno, R.; Chowdhry, S.; Cabello, P.; Isono, K.; Koseki, H.; Honda, T.; Hayes, J.D.; Dinkova-Kostova, A.T.; et al. Crosstalk between NRF2 and HIPK2 shapes cytoprotective responses. Oncogene 2017, 36, 6204–6212.
- Knatko, E.V.; Tatham, M.H.; Zhang, Y.; Castro, C.; Higgins, M.; Dayalan Naidu, S.; Leonardi, C.; de la Vega, L.; Honda, T.; Griffin, J.L.; et al. Downregulation of Keap1 Confers Features of a Fasted Metabolic State. iScience 2020, 23, 101638.
- Knatko, E.V.; Ibbotson, S.H.; Zhang, Y.; Higgins, M.; Fahey, J.W.; Talalay, P.; Dawe, R.S.; Ferguson, J.; Huang, J.T.; Clarke, R.; et al. Nrf2 Activation Protects against Solar-Simulated Ultraviolet Radiation in Mice and Humans. Cancer Prev. Res. 2015, 8, 475–486.
- Taguchi, K.; Maher, J.M.; Suzuki, T.; Kawatani, Y.; Motohashi, H.; Yamamoto, M. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol. Cell. Biol. 2010, 30, 3016–3026.
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109.
More
