Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Vicky Zhou and Version 4 by Vicky Zhou.
SCF (Skp1-Cullin 1-F-box) ligases, which are E3 ubiquitin multi-protein enzymes, catalyse protein ubiquitination and thus allow protein degradation mediated by the 26S proteasome. They play a crucial role in the degradation of cell cycle regulators, regulation of the DNA repair and centrosome cycle and play an important role in several diseases. SCF ligases seem to be needed during all phases of development, from oocyte formation through fertilization, activation of the embryonic genome to embryo implantation.
- ubiquitin-proteasome system
- ubiquitin
- SCF ligases
- oogenesis
- embryogenesis
1. Introduction
Mammalian oogenesis and embryogenesis are extremely important processes. Primordial germ cells are converted into oogonia and subsequently to oocytes, and the diploid cell is transformed into a haploid oocyte ready for fertilization. The oocyte grows and accumulates organelles, mRNA and proteins, which are used during the first embryonic stages [1]. During oogenesis, the oocyte is arrested at the diplotene stage of prophase meiosis I in a structure called a germinal vesicle (GV). Just before ovulation, after the increase in gonadotropin hormones and activation of cyclin dependent kinase (CDK1, a catalytic subunit of the M phase-promoting factor MPF), meiosis is reactivated and the nuclear membrane breaks in a process called germinal vesicle breakdown (GVBD). Subsequently, cytokinesis occurs and the first polar body is extruded. The oocyte enters the second meiotic division and arrest in metaphase II (MII), where the oocyte remains until its fertilization [2]. Only matured oocytes are able to undergo fertilization. Contact with sperm causes intracellular calcium ion oscillation, leading to oocyte activation and meiosis initiation [3]. During fertilization, the haploid oocyte and haploid sperm fuse together to create a zygote, and the first mitotic division occurs within several hours [4]. Early preimplantation development is a highly complicated and strictly regulated process. Preimplantation development starts with several rapid cell cycles with short or completely lacking G phases [5]. These early stages of development are controlled by maternal mRNAs and proteins accumulated during oogenesis. Subsequently, the control of development passes from maternal to embryonic in a process called the maternal-to-zygotic transition (MZT), leading to embryonic genome activation (EGA) [6]. A minor wave of EGA shortly after fertilization is followed by a more robust major wave later in development. The first transcripts were found only 7 h after pronuclei formation in mice [7]. The latest studies suggest that embryonic genome activation is a gradual process that appear with smaller waves of transcription. For example, splicing factor arginine/serine-rich 3 (SRFS3) was found to be already expressed at the 4-cell stage, thus during the minor genome activation in cattle [8]. Maternal reserves are gradually replaced by their embryonic forms and removed from the embryo. Maternal mRNAs are presumably degraded by miRNAs and probably also by other classes of small non-coding RNAs in early embryos [9]. One of the potential pathways of maternal protein degradation is via the ubiquitin-proteasome system (UPS). UPS plays a crucial role in many steps of gametogenesis and embryogenesis.
UPS mediates the proteolysis of a variety of proteins that are important for many basic cellular processes, which include the regulation of cell cycle and development, response to stress, DNA repair, modulation of surface receptors and channels, regulation of transcription and many others. This wide range of processes is due to the large number of individual enzymes that belong to the UPS, which affect a huge number of target substrates. The degradation of proteins by the UPS is based on labelling the targeted protein with ubiquitin. This process is called ubiquitination and involves three enzymatic complexes: E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligases (Figure 1). The E3 enzyme mediates the final interaction between ubiquitin and the substrate, and hence is responsible for substrate specificity. Proteins are polyubiquitinated (labelled with more than just one molecule of ubiquitin) and subsequently directed to degradation by the 26S proteasome [10]. Based on the presence of characteristic domains and mechanisms of ubiquitin transfer from E2 enzyme to the specific substrate proteins, E3 ligases are divided to the three main groups: RING (really interesting new gene) E3s, HECT (homologous to the E6AP carboxyl terminus) E3s and RBR (RING-betweenRING-RING) E3s [11]. One of the most abundant and common RING E3 enzymes is SCF (Skp1-Cullin 1-F-box) ligase, composed of three invariant members: cullin1, SKP1, RBX1 and one of the F-box proteins, which determines the substrate specificity (Figure 2).
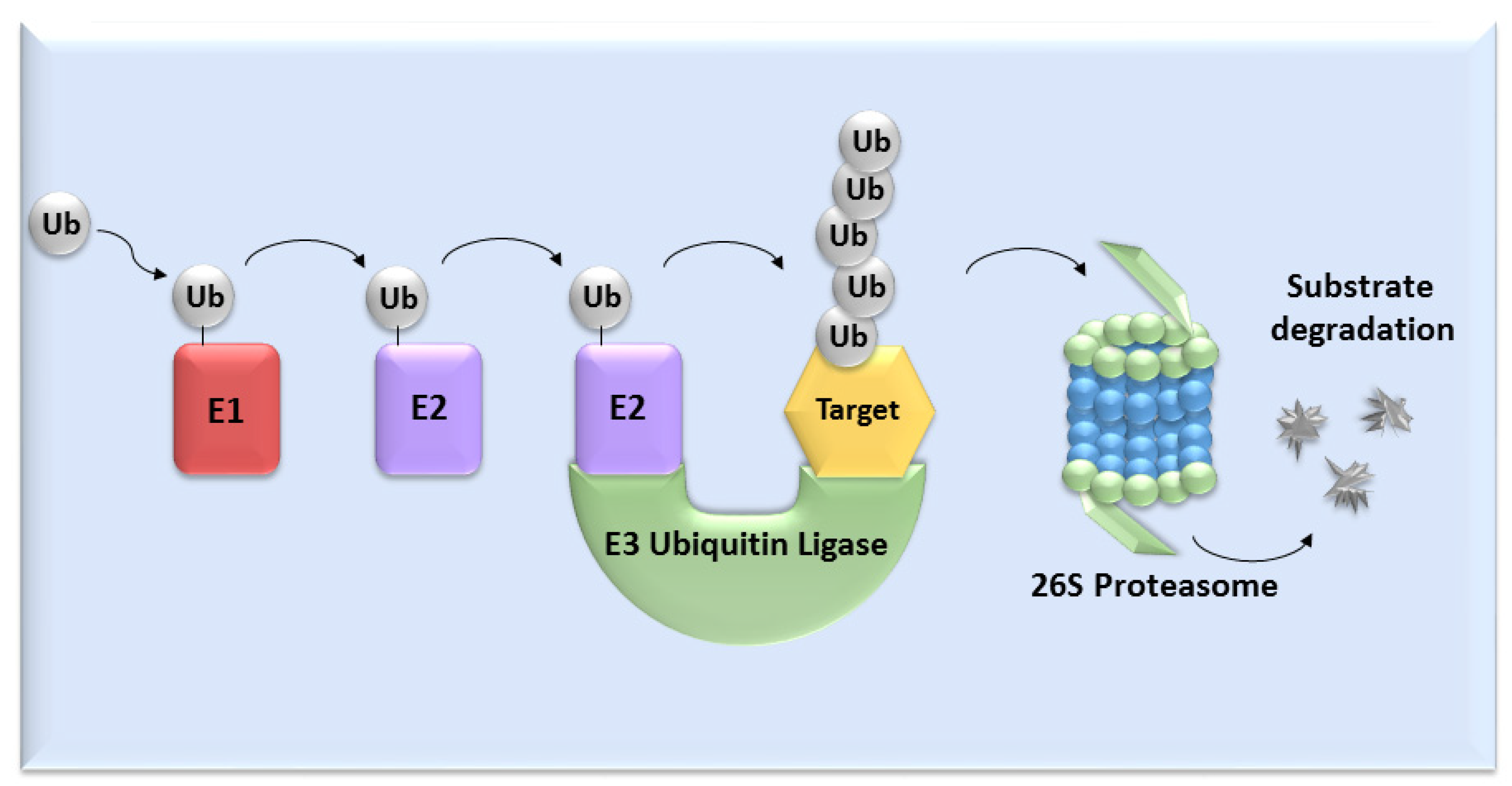
Figure 1. The ubiquitin-proteasome system. Activation and binding of ubiquitin to the target substrate is mediated by three enzymatic complexes (E1, E2 and E3 enzyme). The substrate is selected by E3 ubiquitin ligase and after tagging by ubiquitin, substrate undergoes degradation mediated by 26S proteasome.
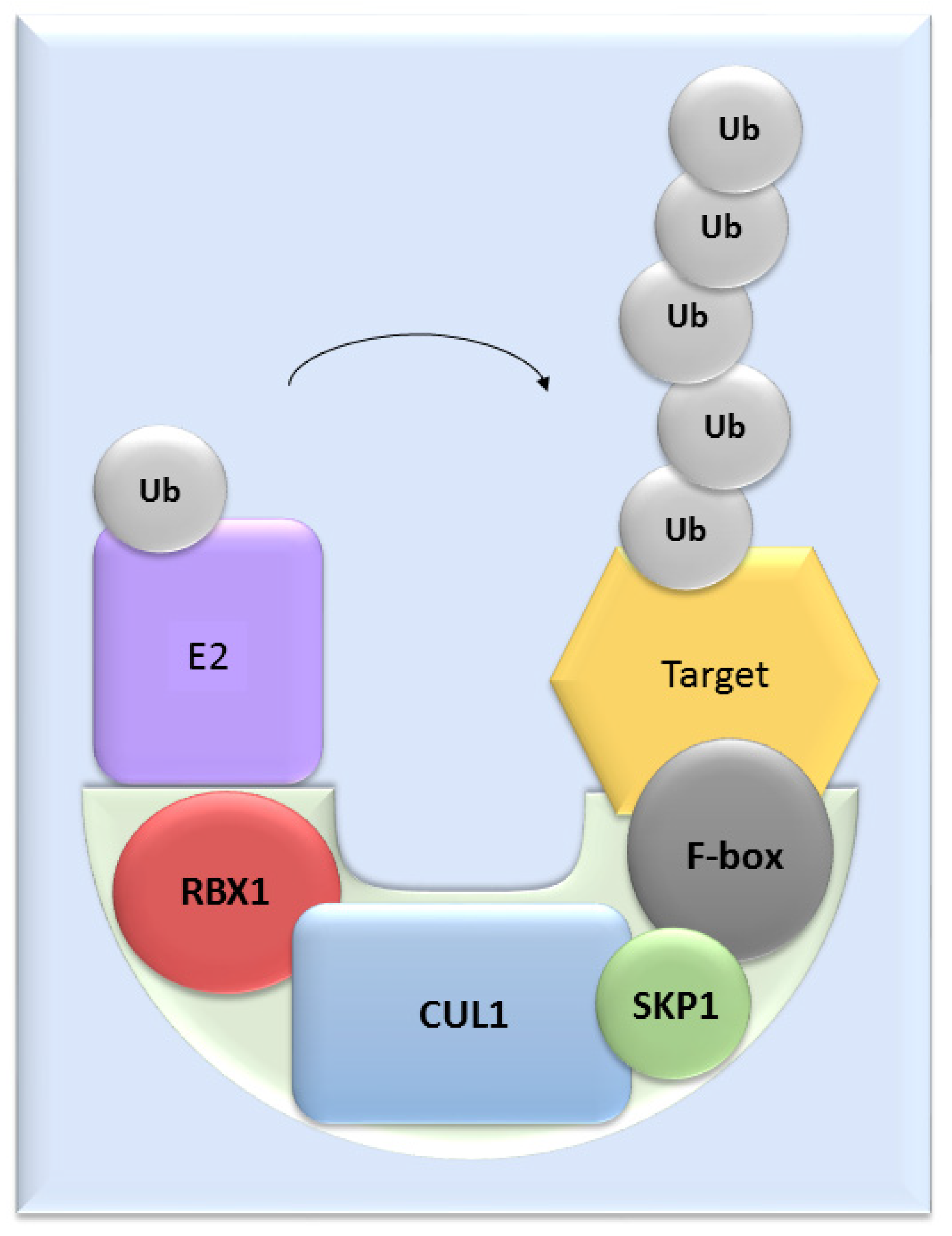
Figure 2. The structure of SCF ligases. One of the most common ligase is composed by RBX1, CUL1, SKP1 protein and a member from the F-box protein family.
2. SCF Ligases Composition
2.1. Cullins
In mammals, seven different cullins are found: Cul1, Cul2, Cul3, Cul4A, Cul4B, Cul5 and Cul7 [12]. The activation of cullins is mediated by neddylation (modification with the protein Nedd8) [13]. After deneddylation, Cul binds to CAND1 (Cullin-associated and neddylation dissociated 1) and inactivates the SCF ligase [14].
In cattle, Cullin1 is expressed from two different genes. The first type is expressed from the MII oocyte until the early 8-cell stage and is called cullin1-like or maternal Cul1. The second one is expressed from the 8-cell stage to the blastocyst stage and is called Cul1 or embryonic Cul1. Both share 83% homology and in somatic cells, the embryonic form of Cul1 is expressed [15]. The expression of Cul1-like mRNA is relatively high from the MII to the 4-cell stage, but at the early 8-cell stage, the mRNA level rapidly decreases and stays low until the blastocyst stage. In contrast, Cul1 mRNA level is low from the MII to the 4-cell stage and gradually increases from the early 8-cell stage, with the highest level at the blastocyst stage [16].
It was recently found that CUL1 is upregulated in lamb oocytes after IVM. This suggests earlier maternal protein degradation, which could contribute to the lower developmental competence of oocytes [17].
2.2. RBX1 (Ring-Box 1)
Rbx1 mediates cullin neddylation and contains a RING finger domain recruiting the E2 enzyme [18]. Rbx1, attached to Cul, forms a catalytic core of SCF ligases and, based on the type of connected cullin, activates several types of SCF ligases [19]. The expression of Rbx1 starts at the time of a major wave of EGA in cattle, at the late 8-cell stage [16]. The silencing of Rbx1 caused embryonic lethality at E7.5 in mouse embryos. This developmental arrest was caused by an accumulation of p27, leading to hypoproliferation [20]. Moreover, the silencing of Rbx1 by RNAi causes defects in the first meiotic division of oocytes and in mitotic chromosomal condensation and segregation, abnormal cortical protrusion, multinucleate cells and defects in germ cell proliferation of Caenorhabditis elegans embryos [21]. The necessity of Rbx1 was also verified by Jia et al. [22], who confirmed that the silencing of Rbx1 induced embryonic lethality of C. elegans. Jia et al. [22] presumed that this could be caused by an accumulation of a number of substrates of SCF ligases, because SCF ligases control the turnover of many short-lived regulatory proteins in a cell content-dependent and spatially dependent manner. Rbx1, namely its homolog called Roc1a, is also required for the cell proliferation and embryonic development of Drosophila melanogaster [23].
2.3. SKP1 (S-Phase Kinase-Associated Protein 1)
Skp1 mediates the binding of the F-box protein to Cul1 [24]. The dimerization of Skp1 causes overlaps of the F-box protein binding site, leading to the prevention of Skp1 and F-box protein interaction [25]. The mRNA transcription of bovine Skp1 starts at the early 8-cell stage, indicating an important role during the EGA that occurs at the late 8-cell stage in cattle [16]. Skp1 also plays an essential role in the prophase I to metaphase I transition during sperm development. SKP1-deficient spermatocytes exhibit precocious pachytene exit, premature desynapsis, loss of PLK1 and BUB1 at centromeres with a persistence of HORMAD, γH2AX, RPA2 and MLH1 in diplonema and reduced MPF activity [26]. Changes in Skp1 expression play a role in several diseases, for example in the development of Parkinson’s disease [27] and lymphomas [28].
2.4. F-Box Proteins
A large number of mammalian F-box proteins have been identified so far. F-box proteins are divided into three subgroups based on their structural conformation: FBLs, FBXs and FBWx. FBW proteins contain WD40-repeat domains, FBL proteins contain leucine-rich repeats and FBX contain other structures [29]. Based on the existence of these motifs, F-box proteins are able to connect a wide range of substrates to the SCF complex. F-box proteins are short-lived proteins regulated by auto-ubiquitination. The result of their auto-ubiquitination is a rapid switching among multiple SCF complexes leading to cells adaptation to changing physiological conditions and progression through individual steps of the cell cycle [30].
3. SCF Ligases in Oocyte Maturation
Recently, more and more studies have been dealing with the role of SCF ligases during oogenesis (Table 1 and Figure 3), and the roles of several SCF ligases have been proven, predominantly in lower organisms [31] or mice [31][32][33]. During oogenesis, oocytes are initially arrested in the prophase of the first meiotic division with a low MPF level. GVBD and progression into metaphase I is mediated by an increasing level of MPF, leading to the first polar body extrusion. Immediately after GVBD, early mitotic inhibitor 1 (Emi1), an inhibitor of anaphase-promoting complex/cyclosome (APC/C), undergoes SCFβTrCP-dependent degradation [34][35]. The degradation of Emi1 is stimulated by phosphorylation and βTrCP binding mediated by Plk1 [36]. APC/C is responsible for cyclin B destruction and thereby inactivation of MPF. Thus, the degradation of Emi1 leads to APC/C activation, subsequent degradation of cyclin B and MPF activation [34][35]. From the MI stage, oocytes continue immediately to the second meiosis. The meiotic arrest of oocytes at the MII stage is driven by the stabilization of the high activity of MPF by cytostatic factor (CSF). CSF inhibits APC/C and thereby prevents the degradation of cyclin B. Another of the CSF components is Emi2, a direct inhibitor of APC/C [37].
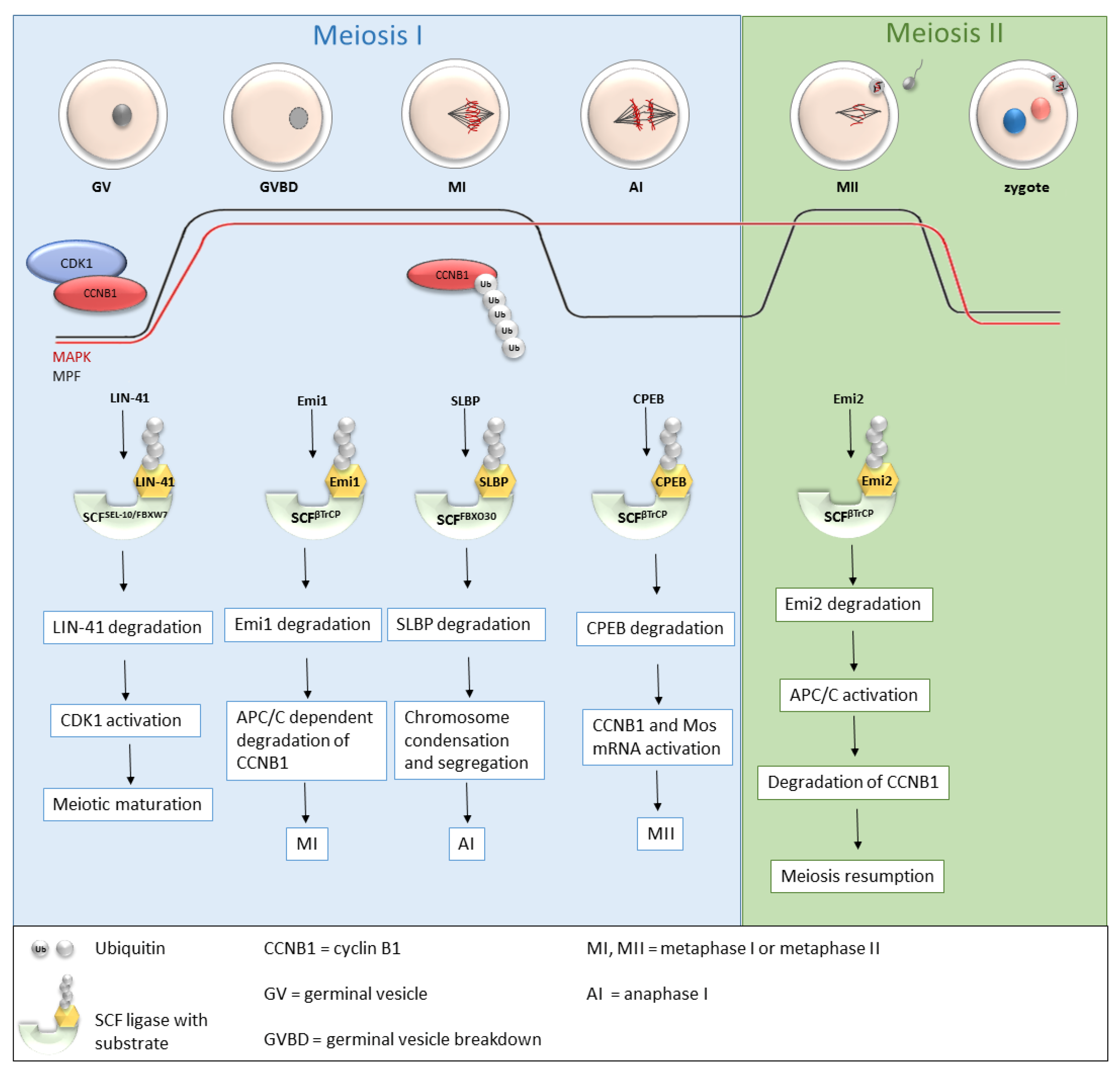

Figure 3. Roles of selected SCF ligases in oogenesis. Progression through oogenesis is mediated by MAPK and MPF (composed by CDK1 and CCNB1) activity which is graphically represented by red and black curves. Activation of CDK1 is regulated by a cyclic activity of CCNB1, whose degradation is mediated via ubiquitination by APC/C. The level of MAPK is stable to the metaphase II. SCFSEL-10/FBXW7, SCFβTrCP and SCFFBXO30-mediated degradation of their targets is essential for progression through meiosis I and meiosis II stages of mouse, C. elegans and Xenopus oocytes. AI—anaphase I, APC/C—anaphase promoting complex/cyclosome, CCNB1—cyclin B1, CDK1—cyclin-dependent kinases I, CPEB—cytoplasmic polyadenylation element-binding protein, Emi1, Emi2—early mitotic inhibitor 1 or 2, GV—germinal vesicle, GVBD—germinal vesicle breakdown, MI, MII—metaphase I, metaphase II, MAPK—mitogen-activated protein kinase, MPF—M phase-promoting factor, SCF—Skp1-Cul1-F-box complex, SLBP—stem-loop-binding protein, Ub—ubiquitin.
Table 1. Known SCF ligases and their substrates in oogenesis of mouse, Xenopus and C. elegans oocytes.
| SCF Ligase | Substrates | Animal | Reference |
|---|---|---|---|
| SCFβTrCP | Emi2/Erp1 | Xenopus laevis | [38] |
| Emi1 | [39] | ||
| CPEB | [40] | ||
| BTG4 | Mouse | [41] | |
| SCFSEL-10/FBXW7/CDC4 | GLD-1,CPB-3 | Caenorhabditis elegans | [31] |
| CDC6 | [42] | ||
| LIN-41 | [43] | ||
| SCFFBXO30 | SLBP | Mouse | [32] |
| SCFFBXO34 | Not identified | Mouse | [33] |
| SCFFBXW15,FBXO12J | Not identified | Mouse | [44] |
4. SCF Ligases in Embryogenesis
Recently, the role of SCF ligases in maternal protein degradation has been frequently discussed [45][46][47][48]. Based on the expression of SCF complex members (Cul1, Skp1, Rbx1) during all stages of preimplantation development, it is presumed that the SCF complex plays an important role during the early embryogenesis of cattle [16][49]. The inhibition of SCF ligases by cultivation in MLN4924 (a specific inhibitor of cullin neddylation) in cattle leads to a statistically significant delay in development and also to a significant increase in the total protein level, although no accumulation of a specific protein has been found yet. Reduced levels of mRNA of EGA markers (PAPOLA, and U2AF1A) were found after MLN4924 treatment [47]. The importance of SCF ligases for oogenesis and embryogenesis is also assumed in mice, since a large number of F-box proteins have been found in oocytes and zygotes. Wang et al. [50] have identified 19 different F-box proteins overexpressed in the mouse oocytes, and presume that these F-box proteins play an important role in protein degradation after fertilization [50]. F-box proteins are also highly abundant in two-cell embryos [51]. The list of SCF ligases and their known substrates important for embryogenesis can be found in Table 2.
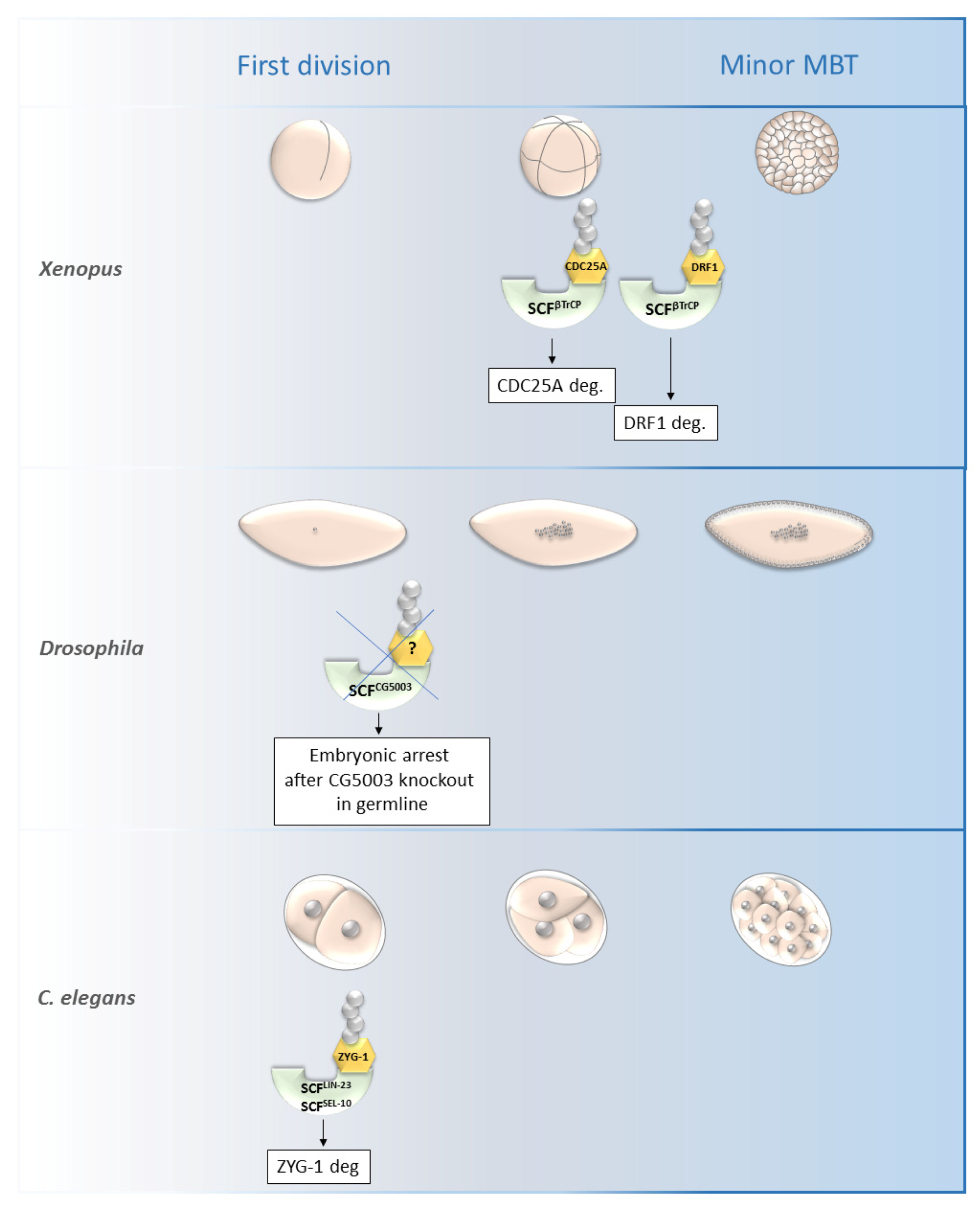
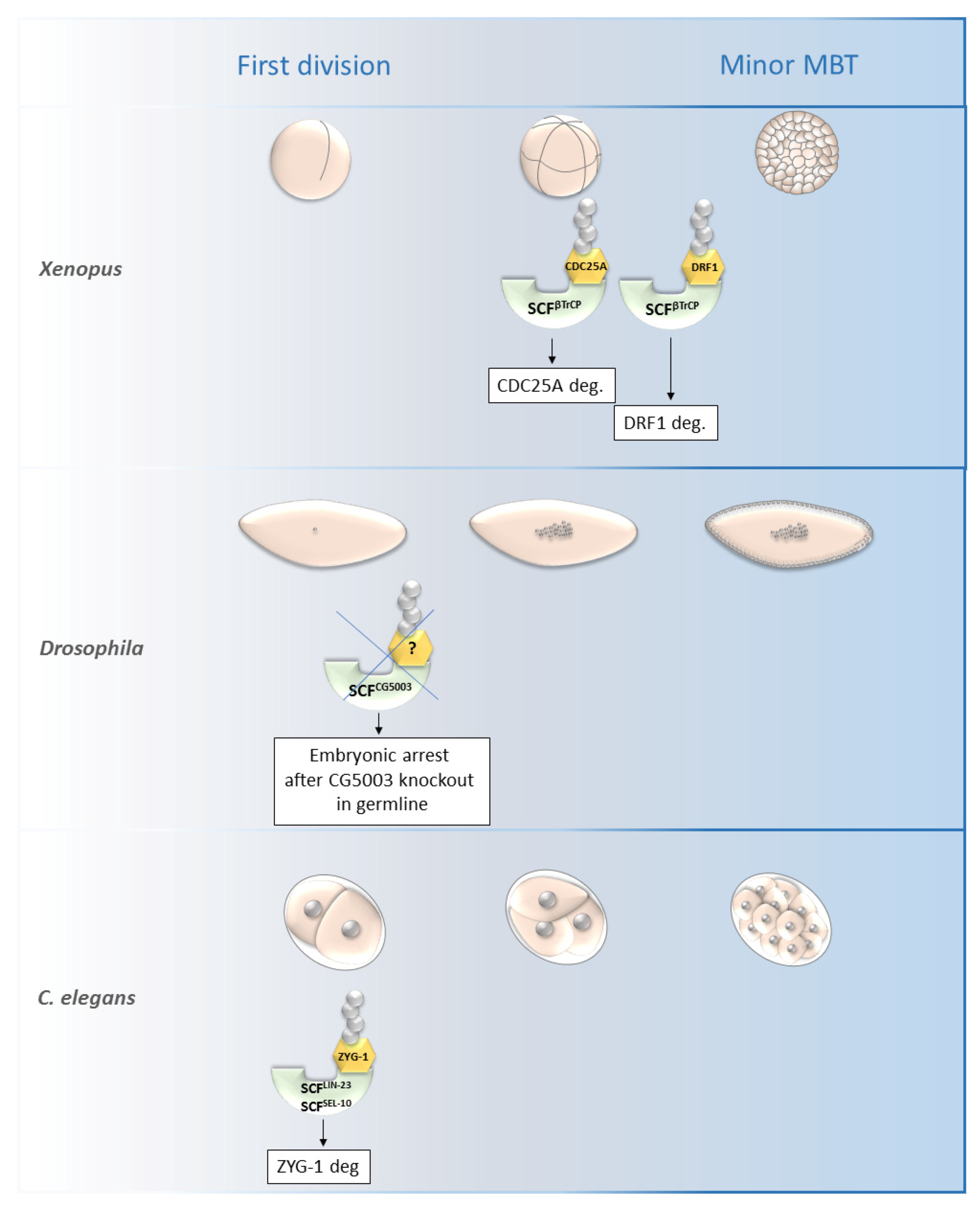 Figure 4. Roles of selected SCF ligases in embryogenesis. Figure shows selected SCF ligases and their roles in degradation of proteins during embryogenesis of Xenopus laevis, Drosophila melanogaster and C. elegans. The main impact of these selected ligases was observed from the first division until the minor MBT. The SCFβTrCP and SCFZYG-1- mediated degradation is important for early embryonic development of Xenopus and C. elegans, respectively. The specific substrate of SCFCG5003 in Drosophila development has not been described yet but its degradation via CG5003 is necessary for progression through the first and second embryonic division. MBT—mid-blastula transition, SCF—Skp1-Cul1-F-box complex, ZYG-1—zygote defective protein, CDC25A—M-phase inducer phosphatase 1.
Figure 4. Roles of selected SCF ligases in embryogenesis. Figure shows selected SCF ligases and their roles in degradation of proteins during embryogenesis of Xenopus laevis, Drosophila melanogaster and C. elegans. The main impact of these selected ligases was observed from the first division until the minor MBT. The SCFβTrCP and SCFZYG-1- mediated degradation is important for early embryonic development of Xenopus and C. elegans, respectively. The specific substrate of SCFCG5003 in Drosophila development has not been described yet but its degradation via CG5003 is necessary for progression through the first and second embryonic division. MBT—mid-blastula transition, SCF—Skp1-Cul1-F-box complex, ZYG-1—zygote defective protein, CDC25A—M-phase inducer phosphatase 1.
Table 2. Known SCF ligases and their substrates in embryogenesis of human, Xenopus, C. elegans, Drosophila and zebrafish embryos.
| SCF Ligases and Their Substrates in Embryogenesis | |||
|---|---|---|---|
| SCF Ligase | Substrates | Animal | Reference |
| SCFβ-TrCP/Slimb/LIN-23 | DRF1 | Xenopus laevis | [46] |
| CDC25A | [45] | ||
| PLK4 | Drosophila melanogaster | [52] | |
| Medea | [53] | ||
| ZYG-1 | Caenorhabditis elegans | [54] | |
| SCFSEL-10/FBXW7/CDC4 | ZYG-1 | Caenorhabditis elegans | [54] |
| SCFFBXO30 | RARγ, BMP | Human | [55] |
| SCFFBXL15 | Smurf1, BMP | Zebrafish | [56] |
| SCFCG5003 | Not identified | Drosophila melanogaster | [57] |
SCFβ-TrCP was found to be necessary for the mid-blastula transition (MBT) in Xenopus as it is responsible for the degradation of DRF1 (a homolog of mammalian DBF4B) (Figure 4).

 Figure 4. Roles of selected SCF ligases in embryogenesis. Figure shows selected SCF ligases and their roles in degradation of proteins during embryogenesis of Xenopus laevis, Drosophila melanogaster and C. elegans. The main impact of these selected ligases was observed from the first division until the minor MBT. The SCFβTrCP and SCFZYG-1- mediated degradation is important for early embryonic development of Xenopus and C. elegans, respectively. The specific substrate of SCFCG5003 in Drosophila development has not been described yet but its degradation via CG5003 is necessary for progression through the first and second embryonic division. MBT—mid-blastula transition, SCF—Skp1-Cul1-F-box complex, ZYG-1—zygote defective protein, CDC25A—M-phase inducer phosphatase 1.
Figure 4. Roles of selected SCF ligases in embryogenesis. Figure shows selected SCF ligases and their roles in degradation of proteins during embryogenesis of Xenopus laevis, Drosophila melanogaster and C. elegans. The main impact of these selected ligases was observed from the first division until the minor MBT. The SCFβTrCP and SCFZYG-1- mediated degradation is important for early embryonic development of Xenopus and C. elegans, respectively. The specific substrate of SCFCG5003 in Drosophila development has not been described yet but its degradation via CG5003 is necessary for progression through the first and second embryonic division. MBT—mid-blastula transition, SCF—Skp1-Cul1-F-box complex, ZYG-1—zygote defective protein, CDC25A—M-phase inducer phosphatase 1.5. Conclusions/Perspectives
Herein summarized current knowledge about the functions of SCF ligases in oogenesis and early embryogenesis. SCFβ-TrCP is one of the best examined ligases in oogenesis and embryogenesis with the most described substrates and functions. It has been demonstrated that SCFβ-TrCP is involved in the regulation of oocyte maturation by the degradation of CPEB, Emi1 and Emi2 [34][39][40]. Their degradation is necessary to overcome meiosis progression and meiosis arrest. During embryogenesis, SCFβ-TrCP plays a pivotal role in the degradation of some maternal proteins, such as DRF1 and CDC25A in Xenopus embryos [45][46]. On the other hand, the protection of BTG4 from β-TrCP-mediated degradation is essential for proper maternal mRNA clearance during mouse development [41]. Another well-known ligase is SCFSEL-10/FBXW7, which regulates the degradation of several RNA-binding proteins during C. elegans oogenesis. These RNA-binding proteins are GLD-1, CPB-3 and LIN-41, which participate in the regulation of gene expression and thus control the progression of meiosis [31][43]. SCFFBXW7 also mediates the degradation of CDC6 protein, which is important for DNA replication, spindle formation and M-phase progression in Xenopus oocytes [42]. Other members of the F-box family have been identified as having functions in oogenesis and embryogenesis but their substrates have not been discovered yet.
The described roles of SCF ligases in meiosis control, chromosome segregation and developmental progression can potentially lead to a better understanding of problems in human assisted reproduction. In the coming years, sciwentists can expect more and more studies to describe new functions of SCF ligases. The balance of protein formation and degradation is obviously key for the proper regulation of cell cycle progression, meiosis and also for the early embryonic development of vertebrates.
References
- Sánchez, F.; Smitz, J. Molecular Control of Oogenesis. Biochim. Biophys. Acta 2012, 1822, 1896–1912.
- Swain, J.E.; Pool, T.B. ART Failure: Oocyte Contributions to Unsuccessful Fertilization. Hum. Reprod. Update 2008, 14, 431–446.
- Kline, D.; Kline, J.T. Repetitive Calcium Transients and the Role of Calcium in Exocytosis and Cell Cycle Activation in the Mouse Egg. Dev. Biol. 1992, 149, 80–89.
- Laurincík, J.; Hyttel, P.; Baran, V.; Eckert, J.; Lucas-Hahn, A.; Pivko, J.; Niemann, H.; Brem, G.; Schellander, K. A Detailed Analysis of Pronucleus Development in Bovine Zygotes in Vitro: Cell-Cycle Chronology and Ultrastructure. Mol. Reprod. Dev. 1998, 50, 192–199.
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic Genome Activation in Vertebrates. Dev. Cell 2017, 42, 316–332.
- Minami, N.; Suzuki, T.; Tsukamoto, S. Zygotic Gene Activation and Maternal Factors in Mammals. J. Reprod. Dev. 2007, 53, 707–715.
- Aoki, F.; Worrad, D.M.; Schultz, R.M. Regulation of Transcriptional Activity during the First and Second Cell Cycles in the Preimplantation Mouse Embryo. Dev. Biol. 1997, 181, 296–307.
- Kanka, J.; Kepková, K.; Nemcová, L. Gene Expression during Minor Genome Activation in Preimplantation Bovine Development. Theriogenology 2009, 72, 572–583.
- Barckmann, B.; Simonelig, M. Control of Maternal MRNA Stability in Germ Cells and Early Embryos. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2013, 1829, 714–724.
- Glickman, M.H.; Ciechanover, A. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev. 2002, 82, 373–428.
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem 2017, 86, 129–157.
- Petroski, M.D.; Deshaies, R.J. Function and Regulation of Cullin-RING Ubiquitin Ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20.
- Jones, J.; Wu, K.; Yang, Y.; Guerrero, C.; Nillegoda, N.; Pan, Z.-Q.; Huang, L. A Targeted Proteomic Analysis of the Ubiquitin-like Modifier Nedd8 and Associated Proteins. J. Proteome Res. 2008, 7, 1274–1287.
- Bennett, E.J.; Rush, J.; Gygi, S.P.; Harper, J.W. Dynamics of Cullin-RING Ubiquitin Ligase Network Revealed by Systematic Quantitative Proteomics. Cell 2010, 143, 951–965.
- Kepkova, K.V.; Vodicka, P.; Toralova, T.; Lopatarova, M.; Cech, S.; Dolezel, R.; Havlicek, V.; Besenfelder, U.; Kuzmany, A.; Sirard, M.-A.; et al. Transcriptomic Analysis of in Vivo and in Vitro Produced Bovine Embryos Revealed a Developmental Change in Cullin 1 Expression during Maternal-to-Embryonic Transition. Theriogenology 2011, 75, 1582–1595.
- Benesova, V.; Kinterova, V.; Kanka, J.; Toralova, T. Characterization of SCF-Complex during Bovine Preimplantation Development. PLoS ONE 2016, 11, e0147096.
- Ying, C.; Yangsheng, W.; Jiapeng, L.; Liqin, W.; Xiaolin, L.; Mingjun, L.; Juncheng, H. Transcriptome Profiles of Pre-Pubertal and Adult in Vitro Matured Ovine Oocytes Obtained from FSH-Stimulated Animals. Reprod. Domest. Anim. 2021, 56, 1085–1094.
- Morimoto, M.; Nishida, T.; Nagayama, Y.; Yasuda, H. Nedd8-Modification of Cul1 Is Promoted by Roc1 as a Nedd8-E3 Ligase and Regulates Its Stability. Biochem. Biophys. Res. Commun. 2003, 301, 392–398.
- Ohta, T.; Michel, J.J.; Schottelius, A.J.; Xiong, Y. ROC1, a Homolog of APC11, Represents a Family of Cullin Partners with an Associated Ubiquitin Ligase Activity. Mol. Cell 1999, 3, 535–541.
- Jia, L.; Sun, Y. RBX1/ROC1-SCF E3 Ubiquitin Ligase Is Required for Mouse Embryogenesis and Cancer Cell Survival. Cell Div. 2009, 4, 16.
- Sasagawa, Y.; Urano, T.; Kohara, Y.; Takahashi, H.; Higashitani, A. Caenorhabditis Elegans RBX1 Is Essential for Meiosis, Mitotic Chromosomal Condensation and Segregation, and Cytokinesis. Genes Cells 2003, 8, 857–872.
- Jia, L.; Bickel, J.S.; Wu, J.; Morgan, M.A.; Li, H.; Yang, J.; Yu, X.; Chan, R.C.; Sun, Y. RBX1 (RING Box Protein 1) E3 Ubiquitin Ligase Is Required for Genomic Integrity by Modulating DNA Replication Licensing Proteins. J. Biol. Chem. 2011, 286, 3379–3386.
- Noureddine, M.A.; Donaldson, T.D.; Thacker, S.A.; Duronio, R.J. Drosophila Roc1a Encodes a RING-H2 Protein with a Unique Function in Processing the Hh Signal Transducer Ci by the SCF E3 Ubiquitin Ligase. Dev. Cell 2002, 2, 757–770.
- Bai, C.; Sen, P.; Hofmann, K.; Ma, L.; Goebl, M.; Harper, J.W.; Elledge, S.J. SKP1 Connects Cell Cycle Regulators to the Ubiquitin Proteolysis Machinery through a Novel Motif, the F-Box. Cell 1996, 86, 263–274.
- Kim, H.W.; Eletsky, A.; Gonzalez, K.J.; van der Wel, H.; Strauch, E.-M.; Prestegard, J.H.; West, C.M. Skp1 Dimerization Conceals Its F-Box Protein Binding Site. Biochemistry 2020, 59, 1527–1536.
- Guan, Y.; Leu, N.A.; Ma, J.; Chmátal, L.; Ruthel, G.; Bloom, J.C.; Lampson, M.A.; Schimenti, J.C.; Luo, M.; Wang, P.J. SKP1 Drives the Prophase I to Metaphase I Transition during Male Meiosis. Sci. Adv. 2020, 6, eaaz2129.
- Mandel, S.A.; Fishman-Jacob, T.; Youdim, M.B.H. Targeting SKP1, an Ubiquitin E3 Ligase Component Found Decreased in Sporadic Parkinson’s Disease. Neurodegener. Dis. 2012, 10, 220–223.
- Piva, R.; Liu, J.; Chiarle, R.; Podda, A.; Pagano, M.; Inghirami, G. In Vivo Interference with Skp1 Function Leads to Genetic Instability and Neoplastic Transformation. Mol. Cell. Biol. 2002, 22, 8375–8387.
- Jackson, P.K.; Eldridge, A.G.; Freed, E.; Furstenthal, L.; Hsu, J.Y.; Kaiser, B.K.; Reimann, J.D. The Lore of the RINGs: Substrate Recognition and Catalysis by Ubiquitin Ligases. Trends Cell Biol. 2000, 10, 429–439.
- Galan, J.M.; Peter, M. Ubiquitin-Dependent Degradation of Multiple F-Box Proteins by an Autocatalytic Mechanism. Proc. Natl. Acad. Sci. USA 1999, 96, 9124–9129.
- Kisielnicka, E.; Minasaki, R.; Eckmann, C.R. MAPK Signaling Couples SCF-Mediated Degradation of Translational Regulators to Oocyte Meiotic Progression. Proc. Natl. Acad. Sci. USA 2018, 115, E2772–E2781.
- Jin, Y.; Yang, M.; Gao, C.; Yue, W.; Liang, X.; Xie, B.; Zhu, X.; Fan, S.; Li, R.; Li, M. Fbxo30 Regulates Chromosome Segregation of Oocyte Meiosis. Cell. Mol. Life Sci. 2019, 76, 2217–2229.
- Zhao, B.-W.; Sun, S.-M.; Xu, K.; Li, Y.-Y.; Lei, W.-L.; Li, L.; Liu, S.-L.; Ouyang, Y.-C.; Sun, Q.-Y.; Wang, Z.-B. FBXO34 Regulates the G2/M Transition and Anaphase Entry in Meiotic Oocytes. Front. Cell Dev. Biol. 2021, 9, 647103.
- Margottin-Goguet, F.; Hsu, J.Y.; Loktev, A.; Hsieh, H.M.; Reimann, J.D.R.; Jackson, P.K. Prophase Destruction of Emi1 by the SCF(BetaTrCP/Slimb) Ubiquitin Ligase Activates the Anaphase Promoting Complex to Allow Progression beyond Prometaphase. Dev. Cell 2003, 4, 813–826.
- Marangos, P.; Verschuren, E.W.; Chen, R.; Jackson, P.K.; Carroll, J. Prophase I Arrest and Progression to Metaphase I in Mouse Oocytes Are Controlled by Emi1-Dependent Regulation of APC(Cdh1). J. Cell Biol. 2007, 176, 65–75.
- Hansen, D.V.; Loktev, A.V.; Ban, K.H.; Jackson, P.K. Plk1 Regulates Activation of the Anaphase Promoting Complex by Phosphorylating and Triggering SCFbetaTrCP-Dependent Destruction of the APC Inhibitor Emi1. Mol. Biol. Cell 2004, 15, 5623–5634.
- Schmidt, A.; Rauh, N.R.; Nigg, E.A.; Mayer, T.U. Cytostatic Factor: An Activity That Puts the Cell Cycle on Hold. J. Cell Sci. 2006, 119, 1213–1218.
- Tung, J.J.; Hansen, D.V.; Ban, K.H.; Loktev, A.V.; Summers, M.K.; Adler, J.R.; Jackson, P.K. A Role for the Anaphase-Promoting Complex Inhibitor Emi2/XErp1, a Homolog of Early Mitotic Inhibitor 1, in Cytostatic Factor Arrest of Xenopus Eggs. Proc. Natl. Acad. Sci. USA 2005, 102, 4318–4323.
- Sako, K.; Suzuki, K.; Isoda, M.; Yoshikai, S.; Senoo, C.; Nakajo, N.; Ohe, M.; Sagata, N. Emi2 Mediates Meiotic MII Arrest by Competitively Inhibiting the Binding of Ube2S to the APC/C. Nat. Commun. 2014, 5, 3667.
- Setoyama, D.; Yamashita, M.; Sagata, N. Mechanism of Degradation of CPEB during Xenopus Oocyte Maturation. Proc. Natl. Acad. Sci. USA 2007, 104, 18001–18006.
- Zhao, L.-W.; Zhu, Y.-Z.; Chen, H.; Wu, Y.-W.; Pi, S.-B.; Chen, L.; Shen, L.; Fan, H.-Y. PABPN1L Mediates Cytoplasmic MRNA Decay as a Placeholder during the Maternal-to-Zygotic Transition. EMBO Rep. 2020, 21, e49956.
- Daldello, E.M.; Le, T.; Poulhe, R.; Jessus, C.; Haccard, O.; Dupré, A. Correction: Control of Cdc6 Accumulation by Cdk1 and MAPK Is Essential for Completion of Oocyte Meiotic Divisions in Xenopus. J. Cell Sci. 2018, 131, jcs215293.
- Spike, C.A.; Huelgas-Morales, G.; Tsukamoto, T.; Greenstein, D. Multiple Mechanisms Inactivate the LIN-41 RNA-Binding Protein To Ensure a Robust Oocyte-to-Embryo Transition in Caenorhabditis Elegans. Genetics 2018, 210, 1011–1037.
- Chesnaye, E.D.L.; Kerr, B.; Paredes, A.; Merchant-Larios, H.; Méndez, J.P.; Ojeda, S.R. Fbxw15/Fbxo12J Is an F-Box Protein-Encoding Gene Selectively Expressed in Oocytes of the Mouse Ovary. Biol. Reprod. 2008, 78, 714–725.
- Shimuta, K.; Nakajo, N.; Uto, K.; Hayano, Y.; Okazaki, K.; Sagata, N. Chk1 Is Activated Transiently and Targets Cdc25A for Degradation at the Xenopus Midblastula Transition. EMBO J. 2002, 21, 3694–3703.
- Collart, C.; Smith, J.C.; Zegerman, P. Chk1 Inhibition of the Replication Factor Drf1 Guarantees Cell-Cycle Elongation at the Xenopus Laevis Mid-Blastula Transition. Dev. Cell 2017, 42, 82–96.e3.
- Kinterova, V.; Kanka, J.; Petruskova, V.; Toralova, T. Inhibition of SCF Complexes during Bovine Oocyte Maturation and Preimplantation Development Leads to Delayed Development of Embryos. Biol. Reprod. 2018, 100, 896–906.
- Toralova, T.; Kinterova, V.; Chmelikova, E.; Kanka, J. The Neglected Part of Early Embryonic Development: Maternal Protein Degradation. Cell. Mol. Life Sci. 2020, 77, 3177–3194.
- Benesova, V.; Kinterova, V.; Kanka, J.; Toralova, T. Potential Involvement of SCF-Complex in Zygotic Genome Activation During Early Bovine Embryo Development. Methods Mol. Biol. 2017, 1605, 245–257.
- Wang, S.; Kou, Z.; Jing, Z.; Zhang, Y.; Guo, X.; Dong, M.; Wilmut, I.; Gao, S. Proteome of Mouse Oocytes at Different Developmental Stages. Proc. Natl. Acad. Sci. USA 2010, 107, 17639–17644.
- Knowles, B.B.; Evsikov, A.V.; de Vries, W.N.; Peaston, A.E.; Solter, D. Molecular Control of the Oocyte to Embryo Transition. Philos. Trans. R. Soc. B Biol. Sci. 2003, 358, 1381–1387.
- Cunha-Ferreira, I.; Bento, I.; Pimenta-Marques, A.; Jana, S.C.; Lince-Faria, M.; Duarte, P.; Borrego-Pinto, J.; Gilberto, S.; Amado, T.; Brito, D.; et al. Regulation of Autophosphorylation Controls PLK4 Self-Destruction and Centriole Number. Curr. Biol. 2013, 23, 2245–2254.
- Muzzopappa, M.; Wappner, P. Multiple Roles of the F-Box Protein Slimb in Drosophila Egg Chamber Development. Development 2005, 132, 2561–2571.
- Peel, N.; Dougherty, M.; Goeres, J.; Liu, Y.; O’Connell, K.F. The C. Elegans F-Box Proteins LIN-23 and SEL-10 Antagonize Centrosome Duplication by Regulating ZYG-1 Levels. J. Cell Sci. 2012, 125, 3535–3544.
- Cheng, X.; Pei, P.; Yu, J.; Zhang, Q.; Li, D.; Xie, X.; Wu, J.; Wang, S.; Zhang, T. F-Box Protein FBXO30 Mediates Retinoic Acid Receptor γ Ubiquitination and Regulates BMP Signaling in Neural Tube Defects. Cell Death Dis. 2019, 10, 551.
- Cui, Y.; He, S.; Xing, C.; Lu, K.; Wang, J.; Xing, G.; Meng, A.; Jia, S.; He, F.; Zhang, L. SCFFBXL15 Regulates BMP Signalling by Directing the Degradation of HECT-Type Ubiquitin Ligase Smurf1. EMBO J. 2011, 30, 2675–2689.
- Avilés-Pagán, E.E.; Kang, A.S.W.; Orr-Weaver, T.L. Identification of New Regulators of the Oocyte-to-Embryo Transition in Drosophila. G3 Genes Genomes Genet. 2020, 10, 2989–2998.
More
