Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 4 by Conner Chen and Version 3 by Conner Chen.
During cell division, the mitotic spindle, a macromolecular structure primarily comprised of microtubules, drives chromosome alignment and partitioning between daughter cells. Mitotic spindles can sense cellular dimensions in order to adapt their length and mass to cell size.
- mitotic spindle
- scaling
- temporal scaling
- cell division
1. Mitotic Spindle Assembly
The mitotic spindle is a bipolar structure mainly composed of microtubules that assembles around chromosomes and orchestrates their equal partitioning between daughter cells (Figure 1). The poles of the mitotic spindle are most of the time organized around two microtubule-organizing centers (MTOCs): the centrosomes in most animal cells and the spindle pole bodies in yeast. At mitotic entry, the interphasic microtubule network is depolymerized and microtubule turnover increases [1][2]. During mitosis, microtubules nucleated at centrosomes [3][4] form two aster-like structures from which emanate astral microtubules directed toward the cell cortex. The two centrosomes are positioned around the rupturing nucleus by a microtubule-dependent process [5]. Concomitantly, a population of microtubules, hereafter termed spindle microtubules, contact and align chromosomes at the metaphase plate. Mitotic spindle assembly largely relies on the ability of microtubules to continuously and stochastically oscillate between phases of polymerization and depolymerization, a process called dynamic instability [6]. The size of the mitotic spindle is directly linked to the properties and the size of each of its individual components, including microtubules, centrosomes and chromosomes (reviewed in [7]). Analyzing how the size and assembly timing of these components is controlled during cell division is thus key to understanding the principles of spatial and temporal scaling of the spindle.
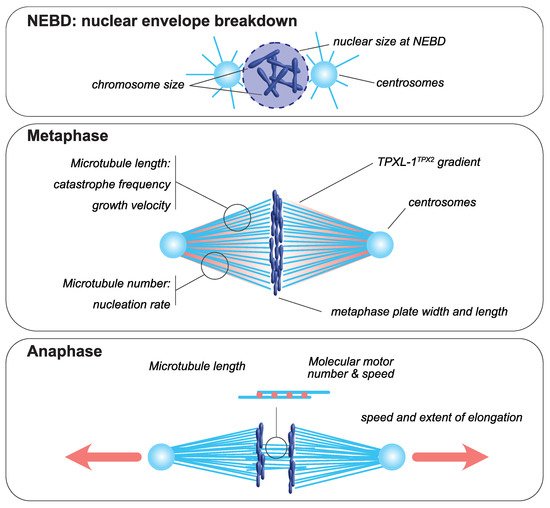
Figure 1. Mitotic spindle components that scale with cell size. At nuclear envelope breakdown, (NEBD, upper panel), chromosome dimensions [8][9] and the size of the nucleus scale with cell size [10][11][12][13][14]. In metaphase (middle panel), the dimensions of the metaphase plate adapt to cellular dimensions [15][9][16]. Centrosome size and a TPXL-1TPX2 gradient scale with cell size [17][18]. The microtubule number, through autocatalytic amplification, scales with the surface area-to-volume ratio to set spindle mass [19]. Spindle microtubule length scales with cell size and is adapted to spindle length [20]. This is achieved by changes in catastrophe frequency [21] and by the scaling of microtubule growth velocity [22][19]. During anaphase (bottom panel), the speed and extent of spindle elongation both scale with cell dimensions [23][24][25].
2. Mechanisms Ensuring Spatial and Temporal Scaling of the Mitotic Spindle
2.1. Microtubule Motors Enable Scaling of Spindle Elongation Rate and Duration during Anaphase
Self-organization of the mitotic spindle is driven by molecular motors that bind and slide along microtubule tracks [26][27][28]. Although microtubule motors can impact mitotic spindle length, their role in spindle length scaling during metaphase has not been reported, and several studies support the idea that molecular motors sliding activity primarily influences spindle shape and organization rather than length [29][30][31][32][33]. This is likely due to the high microtubule turnover in metaphase. During this stage, newly formed microtubules disassemble before associated motors can “sense” their length or the cell volume [34][35]. During anaphase, however, the microtubule turnover is reduced down to dynamics that become compatible with motor-dependent microtubule-length-sensing mechanisms [36][37]. In Schizosaccharomyces pombe, for example, the velocity of spindle elongation during anaphase is proportional to the amount of kinesin-6 Klp9, which increases with cell size [24]. This velocity indeed depends on the number of Klp9 motors bound to midzone microtubules, which is itself directly proportional to the amount of available motors and to the density and length of midzone microtubules. In a feedback mechanism, Klp9 also acts on midzone microtubule length by tuning their growth speed [25]. This makes the duration of spindle elongation and of mitosis constant in this system, and importantly, independent of initial spindle length. Therefore, Klp9 coordinates midzone microtubule sliding and elongation in a cell size-dependent manner, which ensures flawless anaphase midzone elongation with a constant duration. Whether a similar motor-dependent midzone elongation scaling mechanism exists in embryos remains to be explored. In C. elegans embryos, the amplitude and rate of spindle elongation during anaphase scale linearly with spindle and cell size [23]. This allows the extent of chromosome separation to be proportional to cell size, which is essential for efficient karyo- and cytokinesis. During C. elegans embryonic cleavages, the durations of mitosis and of mitotic spindle assembly are short and constant [22][38][39]. Thus, it is likely that the duration of anaphase is also constant throughout early embryonic divisions in this system. The scaling of spindle elongation rate in C. elegans embryos could, as in S. pombe mitosis, contribute to the temporal control of anaphase by maintaining anaphase duration independently of cell size. However, in contrast to S. pombe, anaphase spindle elongation in C. elegans does not primarily rely on midzone microtubule sliding, but rather on cortical pulling forces that are generated on astral microtubules by cortically-anchored dynein motors [40][41][42][43][44]. The amplitude of these pulling forces is proportional to the number of active force generators at the cell cortex, and to the number of astral microtubules contacting the cortex and/or to the surface area contacted by astral microtubules [40][43][45][46]. Conservation of this mechanism outside of C. elegans and nematodes remains to be tested, especially since, in larger embryos, astral microtubules do not necessarily reach the cortex [47]. Alternatively, pulling forces can also be exerted within the cytoplasm without any contact between microtubules and the cell cortex [48][49][50][51][52][53]. Recent in vitro assays reconstituting bulk microtubule motility have demonstrated that cytoplasmic pulling can indeed generate forces, and that the force and velocity of the movement are directly impacted by microtubule length [54]. In either scenario (cytoplasmic versus cortical pulling), the force amplitude depends on the microtubule length [55][56][57] and thus on microtubule dynamics [22][6][58][59].
2.2. Microtubule Dynamics Control Spindle Scaling in Space and in Time
Microtubule dynamics are generally characterized by four parameters: growth and shrinkage velocities and the frequencies of the transition between phases of growth and shrinkage, called catastrophe and rescue, respectively. These four dynamics parameters are sufficient to describe the behavior of a microtubule population in a given context [2][60]. During the cell cycle, microtubule dynamics change [36][37][61] in response to kinase activity [59][62]. In mitosis, microtubules emanate primarily from the two centrosomes and their dynamic properties change dramatically when compared to in interphase [36][62]. This drastic switch in microtubule dynamics sets an optimal average microtubule length around centrosomes, allowing for rapid and efficient chromosome capture [59]. In this high dynamics regime, called “bounded regime”, microtubule length is limited by intrinsic dynamic properties and is particularly sensitive to changes in microtubule growth velocity [59]. Therefore, modulation of microtubule dynamic properties, and thus of microtubule length, represents an efficient mechanism for controlling spindle length and the duration of assembly [22][31][63].
2.2.1. Catastrophe
One of the remarkable changes in microtubule dynamics at mitotic entry is the increase in catastrophe frequency [36][59][61]. This reduction in microtubule lifetime affects microtubule length [59], and, based on in silico models, can also modulate the time of chromosome capture [63]. Thus, scaling of catastrophe frequency with cell size could represent an efficient way of controlling both size and assembly duration of the mitotic spindle. In agreement with this view, an increase in microtubule catastrophe frequency between Xenopus extracts prepared from stage three (four cells) and stage eight (blastula, ~4000 cells) embryos [21] was hypothesized to account for spindle length scaling in cleaving Xenopus embryos. The molecular mechanism proposed to control catastrophe involves a surface-area-sensing mechanism. The increase in the surface area-to-volume ratio, as cell size decreases during successive cleavages, would lead to progressive cortical sequestration of the transport factor Importin-α, through its ability to be anchored to plasma membranes. Cytosolic Importin-α can sequester and inhibit the microtubule-depolymerizing kinesin and catastrophe-inducing factor kif2a. Therefore, its progressive cortical sequestration, as cells get smaller, would in turn allow the release of kif2a into the cytosol and thus the progressive increase in the catastrophe rate [21][64]. This potential mechanism for spindle length scaling does not, however, seem to be conserved among vertebrates, and, in particular, not in zebrafish embryos or in encapsulated Xenopus egg extracts, where the microtubule lifetime does not vary significantly across cleavage [19]. Furthermore, alternative explanations should be considered when analyzing this result. First, caution is required when comparing extracts made from such distant stage embryos. In very large cells, mitotic spindles reach an upper-limit above which spindle length remains almost constant (Figure 2). This is the case for X. laevis mitotic spindle length, which is uncoupled from blastomere size during the first four embryonic divisions [65]. Then, below a given cell diameter of around 140 µm, a feature that seems astonishingly conserved across evolution [66], mitotic spindle length starts scaling linearly with cell size. This feature of spindle length scaling gave rise to the definition of two distinct regimes: the large-cell regime in which mitotic spindle length reaches a plateau and is uncoupled form cell size, and the small-cell regime of linear spindle length scaling [66][67]. Stage three and stage eight Xenopus embryos correspond respectively to the large- and small-cell regimes. Thus, whether the change in catastrophe frequency observed between these two stage extracts underlines mitotic spindle length scaling, or rather represents a feature of the transition point between the large- and the small-cell regimes (Figure 2), remains to be determined. Second, astral and spindle microtubule dynamic properties are distinct and vary independently across embryo cleavage [22][37]. In C. elegans embryos, astral, but not spindle, microtubule catastrophe frequency increases as cells get smaller [22]. The increase in catastrophe frequency between stage three and stage eight Xenopus egg extracts was not measured in spindles per se, but in microtubule asters nucleated from purified human centrosomes introduced in these extracts. Therefore, and potentially in line with the C. elegans in vivo measurements, this experimental context could highlight the behavior of astral, rather than spindle, microtubules. In this later scenario, the variation of catastrophe frequency measured in stage three and stage eight Xenopus embryo extracts is unlikely to affect spindle length. Indeed, a combination of experimental in vivo data in C. elegans and of an in silico model provided evidence that mitotic spindle length scaling is independent of astral microtubule dynamics [22][31]. Finally, an in silico model of spindle assembly [22] predicted that spindle length scaling can be recapitulated by progressively increasing catastrophe frequency, but this was accompanied by a proportional lengthening of the duration of spindle assembly as cells get smaller ([22] and our unpublished data). This result is inconsistent with the observation that mitosis duration is constant across cleavage in different species embryos. Therefore, the potential link between catastrophe frequency, mitotic spindle length scaling and assembly duration in cleaving embryos requires further investigation to specifically address its role in embryos of various size, and in both the large- and small-cell regimes.
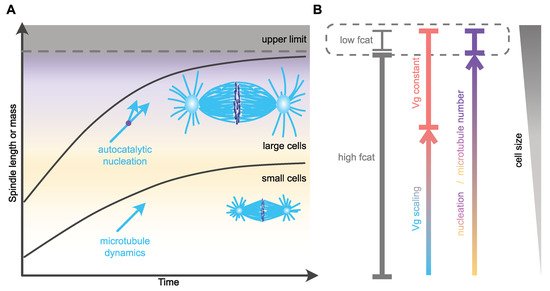
Figure 2. Mechanisms of spatial and temporal control of spindle assembly and scaling. (A) In large embryos, spindle size reaches an upper limit and does not scale with cell size [66][65]. As cell size decreases, spindle size adapts to cell dimensions through the regulation of microtubule nucleation [19]. This mechanism might allow cells to maintain the duration of spindle assembly constant. In small cells, microtubule dynamics and especially growth velocity scales with cell size and regulates microtubule and spindle length [22][19]. In C. elegans, this mechanism allows for the duration of spindle assembly to be constant and independent of cell size [22]. (B) Potential regulation of distinct microtubule parameters with cell size. The variation in catastrophe frequency (fcat) is based on observations made in Xenopus stage 3 and stage 8 embryo extracts, representing large- and small-cell regimes, respectively [21]. Microtubule nucleation scales over a wide range of sizes [19]. In small cells, microtubule growth velocity (Vg) scales with cell size and becomes constant in larger cells [22][19].
2.2.2. Growth Rate
Another microtubule dynamics parameter that can have a profound impact on spindle length in Xenopus egg extracts is the microtubule growth rate. Indeed, progressively raising microtubule growth velocity by adding increasing amounts of the microtubule polymerase XMAP215 in extracts leads to a proportional increase in mitotic spindle length [20]. Similar results are obtained after microinjection of XMAP215 in Xenopus eggs, suggesting that microtubule growth velocity could also regulate spindle length and spindle size scaling in vivo [68]. Although XMAP215 could also act on microtubule nucleation in these experiments (discussed below), modulation of the microtubule growth rate is thus a potential candidate mechanism for the regulation of microtubule and spindle length [69][70][71][72][73][74]. In agreement with this, in the nematode C. elegans and in the sea urchin Paracentrotus lividus microtubule growth rate scales with cell volume during the first rounds of embryonic cleavages [22]. In C. elegans, this correlation between the microtubule growth velocity and cell size does not only promote the length regulation of microtubules and mitotic spindles, it also allows for the duration of mitotic spindle assembly to remain constant and independent of cell and spindle size during cleavage. With mitotic spindle assembly duration being constant in this system, in large blastomeres the spindle assembly rate is higher than in smaller blastomeres, and linearly correlates with the average growth velocity of microtubules [22]. Thus, scaling of the microtubule growth rate with cell volume in embryos appears to be an efficient mechanism for coordinating spatial and temporal scaling of the mitotic spindle during embryonic cleavages. The mechanism by which microtubules can sense cellular volume and modulate their assembly rate accordingly is still unknown, but the limiting component model seems particularly suited [75][76][77]. According to this model, the progressive titration of positive regulators of microtubule assembly by the number of growing microtubule plus-ends could drive the proportional relationship between cell volume, microtubule growth rate and spindle length [22][68].
References
- Saxton, W.M.; Stemple, D.L.; Leslie, R.J.; Salmon, E.D.; Zavortink, M.; McIntosh, J.R. Tubulin Dynamics in Cultured Mammalian Cells. J. Cell Biol. 1984, 99, 2175–2186.
- Walker, R.A.; O’Brien, E.T.; Pryer, N.K.; Soboeiro, M.F.; Voter, W.A.; Erickson, H.P.; Salmon, E.D. Dynamic Instability of Individual Microtubules Analyzed by Video Light Microscopy: Rate Constants and Transition Frequencies. J. Cell Biol. 1988, 107, 1437–1448.
- Kuriyama, R.; Borisy, G.G. Microtubule-Nucleating Activity of Centrosomes in Chinese Hamster Ovary Cells Is Independent of the Centriole Cycle but Coupled to the Mitotic Cycle. J. Cell Biol. 1981, 91, 822–826.
- Vorobjev, I.A.; YuS, C. Centrioles in the Cell Cycle. I. Epithelial Cells. J. Cell Biol. 1982, 93, 938–949.
- Piel, M.; Meyer, P.; Khodjakov, A.; Rieder, C.L.; Bornens, M. The Respective Contributions of the Mother and Daughter Centrioles to Centrosome Activity and Behavior in Vertebrate Cells. J. Cell Biol. 2000, 149, 317–330.
- Mitchison, T.; Kirschner, M. Dynamic Instability of Microtubule Growth. Nature 1984, 312, 237–242.
- Guilloux, G.; Gibeaux, R. Mechanisms of Spindle Assembly and Size Control. Biol. Cell 2020, 112, 369–382.
- Kieserman, E.K.; Heald, R. Mitotic Chromosome Size Scaling in Xenopus. Cell Cycle 2011, 10, 3863–3870.
- Ladouceur, A.M.; Dorn, J.F.; Maddox, P.S. Mitotic Chromosome Length Scales in Response to Both Cell and Nuclear Size. J. Cell Biol. 2015, 209, 645–651.
- Conklin, E. Cell Size and Nuclear Size. J. Exp. Embryol. 1912, 12, 1–98.
- Levy, D.L.; Heald, R. Nuclear Size Is Regulated by Importin Alpha and Ntf2 in Xenopus. Cell 2010, 143, 288–298.
- Levy, D.L.; Heald, R. Mechanisms of Intracellular Scaling. Annu. Rev. Cell Dev. Biol. 2012, 28, 113–135.
- Vuković, L.D.; Jevtić, P.; Edens, L.J.; Levy, D.L. New Insights into Mechanisms and Functions of Nuclear Size Regulation. Int. Rev. Cell Mol. Biol. 2016, 322, 1–59.
- Wilson, E.B. The Karyoplasmic Ratio. In The Cell in Development and Heredity; The Macmillan Company: New York, NY, USA, 1925; pp. 727–733.
- Hara, Y.; Kimura, A. An Allometric Relationship between Mitotic Spindle Width, Spindle Length, and Ploidy in Caenorhabditis Elegans Embryos. Mol. Biol. Cell 2013, 24, 1411–1419.
- Kletter, T.; Reusch, S.; Cavazza, T.; Dempewolf, N.; Tischer, C.; Reber, S. Volumetric Morphometry Reveals Spindle Width as the Best Predictor of Mammalian Spindle Scaling. J. Cell Biol. 2021, 221, e202106170.
- Greenan, G.; Brangwynne, C.P.; Jaensch, S.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Centrosome Size Sets Mitotic Spindle Length in Caenorhabditis Elegans Embryos. Curr. Biol. CB 2010, 20, 353–358.
- Decker, M.; Jaensch, S.; Pozniakovsky, A.; Zinke, A.; O’Connell, K.F.; Zachariae, W.; Myers, E.; Hyman, A.A. Limiting Amounts of Centrosome Material Set Centrosome Size in C. Elegans Embryos. Curr. Biol. 2011, 21, 1259–1267.
- Rieckhoff, E.M.; Berndt, F.; Elsner, M.; Golfier, S.; Decker, F.; Ishihara, K.; Brugués, J. Spindle Scaling Is Governed by Cell Boundary Regulation of Microtubule Nucleation. Curr. Biol. 2020, 30, 4973–4983.e10.
- Reber, S.B.; Baumgart, J.; Widlund, P.O.; Pozniakovsky, A.; Howard, J.; Hyman, A.A.; Julicher, F. XMAP215 Activity Sets Spindle Length by Controlling the Total Mass of Spindle Microtubules. Nat. Cell Biol. 2013, 15, 1116–1122.
- Wilbur, J.D.; Heald, R. Mitotic Spindle Scaling during Xenopus Development by Kif2a and Importin Alpha. Elife 2013, 2, e00290.
- Lacroix, B.; Letort, G.; Pitayu, L.; Salle, J.; Stefanutti, M.; Maton, G.; Ladouceur, A.M.; Canman, J.C.; Maddox, P.S.; Maddox, A.S.; et al. Microtubule Dynamics Scale with Cell Size to Set Spindle Length and Assembly Timing. Dev. Cell 2018, 45, 496–511.e6.
- Hara, Y.; Kimura, A. Cell-Size-Dependent Spindle Elongation in the Caenorhabditis Elegans Early Embryo. Curr. Biol. 2009, 19, 1549–1554.
- Kruger, L.K.; Sanchez, J.L.; Paoletti, A.; Tran, P.T. Kinesin-6 Regulates Cell-Size-Dependent Spindle Elongation Velocity to Keep Mitosis Duration Constant in Fission Yeast. Elife 2019, 8, e42182.
- Krüger, L.K.; Gélin, M.; Ji, L.; Kikuti, C.; Houdusse, A.; Théry, M.; Blanchoin, L.; Tran, P.T. Kinesin-6 Klp9 Orchestrates Spindle Elongation by Regulating Microtubule Sliding and Growth. Elife 2021, 10, e67489.
- Forth, S.; Kapoor, T.M. The Mechanics of Microtubule Networks in Cell Division. J. Cell Biol. 2017, 216, 1525–1531.
- Nazockdast, E.; Redemann, S. Mechanics of the Spindle Apparatus. Semin. Cell Dev. Biol. 2020, 107, 91–102.
- Nedelec, F.J.; Surrey, T.; Maggs, A.C.; Leibler, S. Self-Organization of Microtubules and Motors. Nature 1997, 389, 305–308.
- Dumont, S.; Mitchison, T.J. Force and Length in the Mitotic Spindle. Curr. Biol. 2009, 19, R749–R761.
- Gallaud, E.; Caous, R.; Pascal, A.; Bazile, F.; Gagne, J.P.; Huet, S.; Poirier, G.G.; Chretien, D.; Richard-Parpaillon, L.; Giet, R. Ensconsin/Map7 Promotes Microtubule Growth and Centrosome Separation in Drosophila Neural Stem Cells. J. Cell Biol. 2014, 204, 1111–1121.
- Goshima, G.; Wollman, R.; Stuurman, N.; Scholey, J.M.; Vale, R.D. Length Control of the Metaphase Spindle. Curr. Biol. 2005, 15, 1979–1988.
- Kwok, B.H.; Yang, J.G.; Kapoor, T.M. The Rate of Bipolar Spindle Assembly Depends on the Microtubule-Gliding Velocity of the Mitotic Kinesin Eg5. Curr. Biol. 2004, 14, 1783–1788.
- Metivier, M.; Monroy, B.Y.; Gallaud, E.; Caous, R.; Pascal, A.; Richard-Parpaillon, L.; Guichet, A.; Ori-McKenney, K.M.; Giet, R. Dual Control of Kinesin-1 Recruitment to Microtubules by Ensconsin in Drosophila Neuroblasts and Oocytes. Development 2019, 146, dev171579.
- Needleman, D.J.; Groen, A.; Ohi, R.; Maresca, T.; Mirny, L.; Mitchison, T. Fast Microtubule Dynamics in Meiotic Spindles Measured by Single Molecule Imaging: Evidence That the Spindle Environment Does Not Stabilize Microtubules. Mol. Biol. Cell 2010, 21, 323–333.
- Petry, S. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 2016, 85, 659–683.
- Belmont, L.D.; Hyman, A.A.; Sawin, K.E.; Mitchison, T.J. Real-Time Visualization of Cell Cycle-Dependent Changes in Microtubule Dynamics in Cytoplasmic Extracts. Cell 1990, 62, 579–589.
- Srayko, M.; Kaya, A.; Stamford, J.; Hyman, A.A. Identification and Characterization of Factors Required for Microtubule Growth and Nucleation in the Early C. Elegans Embryo. Dev. Cell 2005, 9, 223–236.
- Arata, Y.; Takagi, H.; Sako, Y.; Sawa, H. Power Law Relationship between Cell Cycle Duration and Cell Volume in the Early Embryonic Development of Caenorhabditis Elegans. Front. Physiol. 2014, 5, 529.
- Galli, M.; Morgan, D.O. Cell Size Determines the Strength of the Spindle Assembly Checkpoint during Embryonic Development. Dev. Cell 2016, 36, 344–352.
- Colombo, K.; Grill, S.W.; Kimple, R.J.; Willard, F.S.; Siderovski, D.P.; Gönczy, P. Translation of Polarity Cues into Asymmetric Spindle Positioning in Caenorhabditis Elegans Embryos. Science 2003, 300, 1957–1961.
- Couwenbergs, C.; Labbé, J.-C.; Goulding, M.; Marty, T.; Bowerman, B.; Gotta, M. Heterotrimeric G Protein Signaling Functions with Dynein to Promote Spindle Positioning in C. Elegans. J. Cell Biol. 2007, 179, 15–22.
- Gotta, M.; Dong, Y.; Peterson, Y.K.; Lanier, S.M.; Ahringer, J. Asymmetrically Distributed C. Elegans Homologs of AGS3/PINS Control Spindle Position in the Early Embryo. Curr. Biol. 2003, 13, 1029–1037.
- Grill, S.W.; Howard, J.; Schäffer, E.; Stelzer, E.H.K.; Hyman, A.A. The Distribution of Active Force Generators Controls Mitotic Spindle Position. Science 2003, 301, 518–521.
- Nguyen-Ngoc, T.; Afshar, K.; Gönczy, P. Coupling of Cortical Dynein and Gα Proteins Mediates Spindle Positioning in Caenorhabditis Elegans. Nat. Cell Biol. 2007, 9, 1294–1302.
- Pecreaux, J.; Röper, J.-C.; Kruse, K.; Jülicher, F.; Hyman, A.A.; Grill, S.W.; Howard, J. Spindle Oscillations during Asymmetric Cell Division Require a Threshold Number of Active Cortical Force Generators. Curr. Biol. 2006, 16, 2111–2122.
- Rodriguez-Garcia, R.; Chesneau, L.; Pastezeur, S.; Roul, J.; Tramier, M.; Pécréaux, J. The Polarity-Induced Force Imbalance in Caenorhabditis Elegans Embryos Is Caused by Asymmetric Binding Rates of Dynein to the Cortex. Mol. Biol. Cell 2018, 29, 3093–3104.
- Howard, J.; Garzon-Coral, C. Physical Limits on the Precision of Mitotic Spindle Positioning by Microtubule Pushing Forces: Mechanics of Mitotic Spindle Positioning. Bioessays 2017, 39, 1700122.
- Gonczy, P.; Schnabel, H.; Kaletta, T.; Amores, A.D.; Hyman, T.; Schnabel, R. Dissection of Cell Division Processes in the One Cell Stage Caenorhabditis Elegans Embryo by Mutational Analysis. J. Cell Biol. 1999, 144, 927–946.
- Grill, S.W.; Hyman, A.A. Spindle Positioning by Cortical Pulling Forces. Dev. Cell 2005, 8, 461–465.
- Kimura, K.; Kimura, A. Intracellular Organelles Mediate Cytoplasmic Pulling Force for Centrosome Centration in the Caenorhabditis Elegans Early Embryo. Proc. Natl. Acad. Sci. USA 2011, 108, 137–142.
- Mitchison, T.J.; Field, C.M. Self-Organization of Cellular Units. Annu. Rev. Cell Dev. Biol. 2021, 37, 23–41.
- Wuhr, M.; Tan, E.S.; Parker, S.K.; Detrich, H.W.; Mitchison, T.J. A Model for Cleavage Plane Determination in Early Amphibian and Fish Embryos. Curr. Biol. 2010, 20, 2040–2045.
- Xie, J.; Minc, N. Cytoskeleton Force Exertion in Bulk Cytoplasm. Front. Cell Dev. Biol. 2020, 8, 69.
- Palenzuela, H.; Lacroix, B.; Sallé, J.; Minami, K.; Shima, T.; Jegou, A.; Romet-Lemonne, G.; Minc, N. In Vitro Reconstitution of Dynein Force Exertion in a Bulk Viscous Medium. Curr. Biol. 2020, 30, 4534–4540.e7.
- Bieling, P.; Telley, I.A.; Surrey, T. A Minimal Midzone Protein Module Controls Formation and Length of Antiparallel Microtubule Overlaps. Cell 2010, 142, 420–432.
- Kimura, A.; Onami, S. Computer Simulations and Image Processing Reveal Length-Dependent Pulling Force as the Primary Mechanism for C. Elegans Male Pronuclear Migration. Dev. Cell 2005, 8, 765–775.
- Minc, N.; Burgess, D.; Chang, F. Influence of Cell Geometry on Division-Plane Positioning. Cell 2011, 144, 414–426.
- Mitchison, T.; Kirschner, M. Microtubule Assembly Nucleated by Isolated Centrosomes. Nature 1984, 312, 232–237.
- Verde, F.; Dogterom, M.; Stelzer, E.; Karsenti, E.; Leibler, S. Control of Microtubule Dynamics and Length by Cyclin A- and Cyclin B-Dependent Kinases in Xenopus Egg Extracts. J. Cell Biol. 1992, 118, 1097–1108.
- Shelden, E.; Wadsworth, P. Observation and Quantification of Individual Microtubule Behavior in Vivo: Microtubule Dynamics Are Cell-Type Specific. J. Cell Biol. 1993, 120, 935–945.
- Rusan, N.M.; Fagerstrom, C.J.; Yvon, A.M.; Wadsworth, P. Cell Cycle-Dependent Changes in Microtubule Dynamics in Living Cells Expressing Green Fluorescent Protein-Alpha Tubulin. Mol. Biol. Cell 2001, 12, 971–980.
- Verde, F.; Labbe, J.C.; Doree, M.; Karsenti, E. Regulation of Microtubule Dynamics by Cdc2 Protein Kinase in Cell-Free Extracts of Xenopus Eggs. Nature 1990, 343, 233–238.
- Wollman, R.; Cytrynbaum, E.N.; Jones, J.T.; Meyer, T.; Scholey, J.M.; Mogilner, A. Efficient Chromosome Capture Requires a Bias in the “search-and-Capture” Process during Mitotic-Spindle Assembly. Curr. Biol. 2005, 15, 828–832.
- Brownlee, C.; Heald, R. Importin Alpha Partitioning to the Plasma Membrane Regulates Intracellular Scaling. Cell 2019, 176, 805–815.e8.
- Wuhr, M.; Chen, Y.; Dumont, S.; Groen, A.C.; Needleman, D.J.; Salic, A.; Mitchison, T.J. Evidence for an Upper Limit to Mitotic Spindle Length. Curr. Biol. CB 2008, 18, 1256–1261.
- Crowder, M.E.; Strzelecka, M.; Wilbur, J.D.; Good, M.C.; von Dassow, G.; Heald, R. A Comparative Analysis of Spindle Morphometrics across Metazoans. Curr. Biol. 2015, 25, 1542–1550.
- Mitchison, T.J.; Ishihara, K.; Nguyen, P.; Wuhr, M. Size Scaling of Microtubule Assemblies in Early Xenopus Embryos. Cold Spring Harb. Perspect Biol. 2015, 7, a019182.
- Milunovic-Jevtic, A.; Jevtic, P.; Levy, D.L.; Gatlin, J.C. In Vivo Mitotic Spindle Scaling Can Be Modulated by Changing the Levels of a Single Protein: The Microtubule Polymerase XMAP215. Mol. Biol. Cell 2018, 29, 1311–1317.
- Flor-Parra, I.; Iglesias-Romero, A.B.; Chang, F. The XMAP215 Ortholog Alp14 Promotes Microtubule Nucleation in Fission Yeast. Curr. Biol. 2018, 28, 1681–1691.e4.
- Gunzelmann, J.; Rüthnick, D.; Lin, T.-C.; Zhang, W.; Neuner, A.; Jäkle, U.; Schiebel, E. The Microtubule Polymerase Stu2 Promotes Oligomerization of the γ-TuSC for Cytoplasmic Microtubule Nucleation. Elife 2018, 7, e39932.
- King, B.R.; Moritz, M.; Kim, H.; Agard, D.A.; Asbury, C.L.; Davis, T.N. XMAP215 and γ-Tubulin Additively Promote Microtubule Nucleation in Purified Solutions. Mol. Biol. Cell 2020, 31, 2187–2194.
- Popov, A.V.; Severin, F.; Karsenti, E. XMAP215 Is Required for the Microtubule-Nucleating Activity of Centrosomes. Curr. Biol. 2002, 12, 1326–1330.
- Roostalu, J.; Cade, N.I.; Surrey, T. Complementary Activities of TPX2 and ChTOG Constitute an Efficient Importin-Regulated Microtubule Nucleation Module. Nat. Cell Biol. 2015, 17, 1422–1434.
- Thawani, A.; Kadzik, R.S.; Petry, S. XMAP215 Is a Microtubule Nucleation Factor That Functions Synergistically with the γ-Tubulin Ring Complex. Nat. Cell Biol. 2018, 20, 575–585.
- Goehring, N.W.; Hyman, A.A. Organelle Growth Control through Limiting Pools of Cytoplasmic Components. Curr. Biol. 2012, 22, R330–R339.
- Good, M.C.; Vahey, M.D.; Skandarajah, A.; Fletcher, D.A.; Heald, R. Cytoplasmic Volume Modulates Spindle Size during Embryogenesis. Science 2013, 342, 856–860.
- Hazel, J.; Krutkramelis, K.; Mooney, P.; Tomschik, M.; Gerow, K.; Oakey, J.; Gatlin, J.C. Changes in Cytoplasmic Volume Are Sufficient to Drive Spindle Scaling. Science 2013, 342, 853–856.
More
