Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Giorgia Imparato.
Organ on chip (OOC) has emerged as a major technological breakthrough and distinct model system revolutionizing biomedical research and drug discovery by recapitulating the crucial structural and functional complexity of human organs in vitro. OOC are rapidly emerging as powerful tools for oncology research. Indeed, Cancer on chip (COC) can ideally reproduce certain key aspects of the tumor microenvironment (TME), such as biochemical gradients and niche factors, dynamic cell–cell and cell–matrix interactions, and complex tissue structures composed of tumor and stromal cells.
- cancer on chip
- tumor microenvironment
1. Organ on Chip Technology Applied in Cancer: Cancer on Chip
OOCs have emerged in the last years as the new frontier in high-throughput screening technology, in drug assessment and development, and in other domains such as nutraceutics, and cosmeceutics identification. OOCs hold the potential to reduce animal testing and provide realistic human cell and tissue in vitro assays [69][1]. The objective of OOC technology is not to build a complete living organ, but to synthesize minimal units that recapitulate the functions at the level of tissues and organs [38][2]. In general, the OOC is provided with culture compartments, often separated by a porous membrane, in which 3D tissues, often consisting of several cell types, can be cultured while microchannels assure nutrient supply. OOC takes advantage of the recent development of microfabrication techniques, and can be “custom-designed” to better mimic tissue-specific function. Through the right choice and the design of materials in the chips and the introduction of electrodes to deliver electrical/mechanical stimuli, it is possible to recapitulate the tissue-specific microenvironment and control the behavior of cells. At the same time, the microchannels provide the cells with the necessary nutrients and remove waste and can be precisely engineered to assure the 3D tissues with the correct (bio)chemical environment, as in the body [57][3]. A sort of classification of OOC can be made by identifying: OOC designed as single channel, compartmentalized, or membrane chips-based system (Figure 21). The complete chips are typically a few cm in size and made up by optically accessible plastic, glass, or flexible polymers [57][3]. In addition to the choice of materials, stimulation, and sensing, the cell sourcing represents a key issue in OOC technology. Cells used in OOC come from three main sources: cell lines, primary cells from human donors, and human induced pluripotent stem cells (hiPSCs). To date, by using hiPSCs, primary cells, and cell line, several tissues and/or organ types have been successfully modeled to reproduce corresponding functional subunits, including, for example, the brain [30][4], heart [70][5], lung [10,25,71][6][7][8], liver [26[9][10],27], intestine [72[11][12][13],73,74], vasculature [75[14][15][16],76,77], kidney [78][17]. Importantly, these OOC devices can reproduce organ level response to exogenous agents such as inflammatory responses of the lung to silica nanoparticles, [25][7] intestinal epithelial-microbiome crosstalk, [73][12] early liver fibrotic activation in response to anti metabolites chemotherapy drug [79][18] as well as flow dependent recruitment of circulating immune cells [80[19][20],81], and organ specific inflammatory reaction in vitro. Moreover, they can also effectively mimic many types of organ specific disease states, including pulmonary oedema and thrombosis, asthma, inflammatory bowel disease, paving the way for a new era in drug development and new therapeutic discovery [16,31][21][22]. As oncology is one of the most important targets of drug discovery, its is in this area that a number of advances in the creation of more physiologically relevant approaches, such as COC, are most evident [16,57,82][21][3][23]. The typology of the cells used to produce COC is similar to that used in OOC. COC often uses cell lines but resulting in inconsistencies between the model and an original tumor. This limitation can be solved with the use of patient biopsies that would generate PDX-tumor on chip models that would represent more powerful models than current ones, avoiding the use of established lines. Regarding the use of iPSC, it has emerged as the most promising candidate for OOCs since it can be produced from almost every type of adult cell, including skin-, blood-, or hair cells, and can be used to produce many different cell types that are present in various organs of the body, and that would otherwise be very difficult to obtain, such as cells from the heart, brain, lung, liver, gut, and also blood vessels. Despite this extensive use in OOC, we still have very few iPSC-based COC. The two most crucial bottlenecks in the establishment of iPSC cancer models are the efficiency of malignant-cell reprogramming and the ability to differentiate iPSCs into the cell type of interest. A few published studies and anecdotal reports suggest that cancer cells are generally more refractory to reprogramming than normal cells [83][24]. Several COC models have been developed in the last years allowing one to manipulate the TME for studying cell behavior under specific metabolic gradients conditions [84][25] or to study the TME changes correlated to CAF interaction and vice versa [16,21,40,85,86][21][26][27][28][29]. Controlled parameters and read-out methods can be different among chip types, but the read-outs are commonly based on cell and invasive lesion tracking, [87][30] gradient sensing, staining, and gene expression quantification using RT-qPCR [39][31]. The COC community has devoted significant attention to visualizing ECM components and remodeling, for which, thank to the optical accessibility of the microfluidic devices, different microscopy and imaging techniques can be used, such as second harmonic generation (SHG), confocal reflectance microscopy and immunofluorescence [39,88,89,90][31][32][33][34]. This enables simultaneous interrogation of ECM composition and structure with the measurement of transport parameters. This is particularly advantageous when cell synthesized matrices are used in the microfluidic devices, since provide a more representative tumor ECM allowing for the quantification of the contribution of each ECM constituent to transport properties [88,91,92][32][35][36]. Some of the earliest applications of microfluidic cell culture technology focused on modeling specific steps in the cancer cascade, including tumor growth and expansion, angiogenesis, progression from early to late stage lesions involving an EMT, tumor cell invasion and metastasis. Here, we concentrate our attention on COC designed for recreating tissue–tissue interfaces, crucial for reconstituting in a physiological context the interaction occurring between tumor and its environment during cancer invasion and metastasis. Owing to several other interesting reviews looking into tumor chips including endothelial and immune cells [17,19,93,94,95][37][38][39][40][41] we do not discuss the tumor–endothelial–immune cells interactions in this paper but mainly debate cancer-cell–ECM interactions during tumor growth and invasion, focusing on the role of biophysical properties of ECM in guiding the pathological tumoral process.
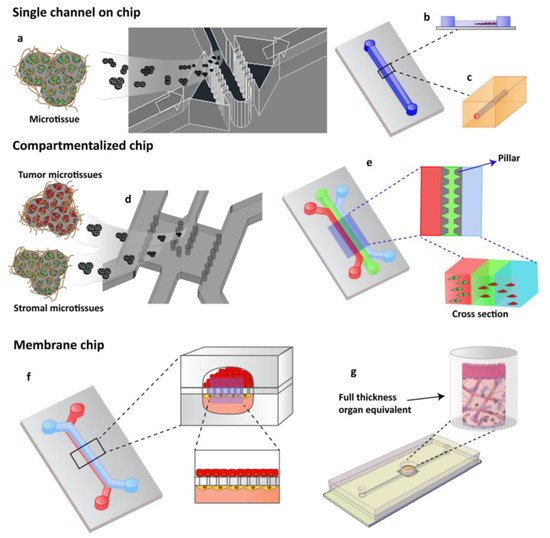
Figure 21. OOCs designs with different cell culture options. (a) Single channels chip designed by Garziano et al. to induce the assembly of dermal microtissues and on-line monitoring the newly synthesized collagen network, by means of SHG imaging, allowing to quantify in real time the effect of perfusion flow and biochemical stimulation on the newly collagen assembly degree. Reproduced from [96][42] with permission from The Royal Society of Chemistry. (b) Lumen channel designed to model human breast cancer cells collective invasion from primary tumors in response to interstitial fluid pressure, as reported by Piotrowski-Daspit et al. [87][30] or (c) designed to mimic a microvessel within a collagen gel scaffold as reported by Pauty et al. to study VEGF-A-induced angiogenesis permeability and angiogenic inhibitors effects [76][15]. (d,e) Compartmentalized chip: In these devices, pillars are used to separate microchannels in which 3D cell culturing is possible. The microchannels are independently addressable in order to fill each channel with a specific cell population and to allow heterotypic cultures. Depending upon the design of the chip, a 3D cell culture can occur by injecting the specific cell populated hydrogel in the correspondent channel or filling the microchamber with specific 3D microtissue. (d) The micropillar guarantees the physical contact of different cellular species embedded in their own ECM [93][39] or (e) in an ECM-like matrix. (f,g) Membrane chip. These devices allow a co-culture in a series of microchannels/chambers separated by a porous membrane. This multi-layered chip type was originally developed to mimic the endo- and epithelial cell layers found in the lung by Ingber [24][43]. Further, different devices have been designed in order to perform 3D organotypic culture by using both (f) membrane or (g) transwell insert.
2. Coupling Cancer on Chip and Artificial Intelligence for Future Cancer Management
Currently, automated systems are utilized in a clinical setting to identify specific target cell types, and the digitization of histopathological images has enabled more precise analyses. The resulting data can be input into machine learning (ML) algorithms for pattern recognition without the need for frequent fine-tuning of parameters or supervision [117][44]. Moreover, the development of high-throughput whole-slide imaging technologies has opened avenues for the ML analysis of immunohistochemical (IHC) source data to yield large datasets for precision oncology [118][45]. Thus, compared with the more traditional approach to IHC pattern analysis at the cellular level, ML models enable a more holistic analysis of the TME at the whole-slide level [119][46]. AI is already being used to analyze various tumor types, such as breast and lung cancers [120][47], and is poised to elevate histological analysis to a more sophisticated level. Given the established strengths of ML techniques in IHC, the potential application of AI in the analysis of tumor models and COC technology is an exciting prospect. Studies using COC have yielded huge quantities of data, and Elmusrati and Ashammakhi [36][48], as well as Fetah et al. [33][49], have proposed the use of ML models to extract and maximize this information. Indeed, the use of COC technology allows for the integration of complex assays and non-invasive real-time monitoring of important cellular and extracellular parameters exploiting for example advanced imaging technologies. Therefore, they represent high-throughput systems that will help to derive sound conclusions on the basis of a massive amount of data. The latter need for appropriate data management and analysis system. Therefore, in parallel to the production of novel COC platforms, ML algorithms to manage these data have been developed. In addition, although traditional ML offers advanced data processing capabilities, the advent of its most important component, the deep learning, made possible to analyze massive unstructured data such as images, drug–target interactions, and computational biology [16,17,18,19,34][21][37][50][38][51]. An example of combination of advanced live cell imaging algorithm and artificial intelligence with COC technology is reported by Oliver et al. [35][52] They have developed a platform for detecting cancer cells with a brain metastatic phenotype combining AI, a blood–brain barrier on a chip and confocal tomography to discern between the metastatic signatures of cancer cells. Using their chip, they typify the migratory and proliferative phenotypes of cancer cells having varying degrees of brain metastatic potential as well as cells from cancer patient samples with known metastatic potential. By combining these results with AI, it is possible to predict the metastatic potential of cancer cells. Recent evidences report that diagnosis based on SHG images and ML can support the rapid and accurate detection of some kinds of cancer in clinical practice [121][53]. COC device hosting engineered tumoral tissue presenting native ECM (cell-synthesized or cell-derived) have been already used to be analyzed by SHG-MPM to detect pathological alterations to the ECM. Therefore, they are a promising platform to implement ML into the SHG image postprocessing to enhance differentiation of normal and tumor tissues, potentially enabling automated screening of tissue in COC under a different therapeutic regime. Nonetheless, the integration of ML algorithms for microfluidic tumor model analysis remains in its infancy, with few demonstrated examples. Several limitations of AI in the analysis of tumor models have to be overcome, including the lack of sufficient, high-quality datasets. Further research is thus warranted, to enable us to harness the full potential of bringing together in vitro models and AI in the study of the TME and to enable us to extract and maximize the vast quantity of information stored within the intricacies of the TME. This translates into improved diagnostic, prognostic, and therapeutic outcomes for patients [122][54].
3. Pros and Cons of Cancer on Chip
There is a wide range of tumor models, each with distinct advantages and disadvantages. Due to the inherent differences in complexity and functionality, the choice of model is usually dependent on the application. For example, compartmentalized COCs replicate the pato-physiological separation existing between cancer mass and stroma allowing to perform advanced invasion assays to see how cancer cells cross the barrier and interact with the stromal population. [59][55] In this perspective, compartmentalized COCs, faithfully recapitulate the TME and cell heterogeneity and allow one to simulate the endothelial network, resulting as being particularly useful in studies focusing on the neovascularization, invasion, and dissemination of cancer cells [38][2]. In general, COCs models are less costly than animals; pre-clinical models do not present ethical concerns and solve the differences that exist between humans and animal species that make the translation of preclinical results from animals to humans not always possible. Despite standard 3D cell culture, COCs present the advantage not only to better recapitulate native human microenvironments, but also to be provided with electrodes and sensors allowing one to model and control mechanical stresses, fluid flow, oxygen levels, temperature, and pH [97][56]. At the same time, however, COCs are more difficult to use than many other 3D culture systems and their use needs highly specialized personnel which can result in increasing experimental costs. However, compared with standard 3D human macroscale models, the addition of the chip allows the obtaining of high-resolution images that allow one to determine where in the tissue to look. Technical robustness is another challenge, as the small scale and complexity of microfluidic systems that experience controlled fluid flow require that many factors must interplay perfectly to achieve optimal functionality, and simple factors, such as bubble formation, can ruin an experiment. For long-term studies, there is the challenge of maintaining cell viability and functionality and structural integrity of multiple tissues and different cell types using a common media and consistent fluid flow [123,124][57][58]. In addition, the use of COCs do not allow one to recreate tumor at real size (typically > 109 cells) but it is possible to reach a cellular range of 106 cells and more importantly, to recapitulate cellular heterogeneity which is a fundamental feature of tumor behavior. At last, it is to be noted that, until now, while extraordinary advances have been attained in integrating a miniaturized tool for controlling and sensing directly on-chip, the design and realization of tissue engineered constructs generally used on the chip is still very poor. The generalized use of exogenous materials as cell scaffolding along with a poor control of microenvironmental condition at single cell level, strongly limit COC current reliability and robustness. The next development in this field aims at designing COC devices able to take into account not only the three-dimensionality of the tissue, but also the time and space molecular presentation and morphophysical features of the cell microenvironment in tissue (patho-) physiology. This is strongly required, particularly, to mimic events in which ECM dynamics play a pivotal role such as the development process, aging, fibrosis, desmoplastic reaction [125][59].
References
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2021, 20, 345–361.
- Ruiz-espigares, J.; Nieto, D.; Moroni, L.; Jiménez, G.; Marchal, J.A. Evolution of Metastasis Study Models toward Metastasis- On-A-Chip: The Ultimate Model? Small 2021, 17, 2006009.
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772.
- Van Der Helm, M.W.; van der Meer, A.; Eijkel, J.C.; van den Berg, A.; Segerink, L.I. Microfluidic organ-on-chip technology for blood-brain barrier research. Tissue Barriers 2016, 4, e1142493.
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.-R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V.; et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016, 110, 45–59.
- Hassell, B.; Goyal, G.; Lee, E.; Sontheimer-Phelps, A.; Levy, O.; Chen, C.; Ingber, D.E. Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Rep. 2017, 21, 508–516.
- Huh, D.; Leslie, D.C.; Matthews, B.D.; Fraser, J.P.; Jurek, S.; Hamilton, G.A.; Thorneloe, K.S.; McAlexander, M.A.; Ingber, D.E. A Human Disease Model of Drug Toxicity–Induced Pulmonary Edema in a Lung-on-a-Chip Microdevice. Sci. Transl. Med. 2012, 4, 159ra147.
- Zamprogno, P.; Wüthrich, S.; Achenbach, S.; Thoma, G.; Stucki, J.D.; Hobi, N.; Schneider-Daum, N.; Lehr, C.M.; Huwer, H.; Geiser, T.; et al. Second-generation lung-on-a-chip with an array of stretchable alveoli made with a biological membrane. Commun. Biol. 2021, 4, 1–10.
- Corrado, B.; De Gregorio, V.; Imparato, G.; Attanasio, C.; Urciuolo, F.; Netti, P.A. A three-dimensional microfluidized liver system to assess hepatic drug metabolism and hepatotoxicity. Biotechnol. Bioeng. 2019, 116, 1152–1163.
- Bhise, N.S.; Manoharan, V.; Massa, S.; Tamayol, A.; Ghaderi, M.; Miscuglio, M.; Lang, Q.; Zhang, Y.S.; Shin, S.R.; Calzone, G.; et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016, 8, 014101.
- Shin, W.; Hinojosa, C.D.; Ingber, D.E.; Kim, H.J. Human Intestinal Morphogenesis Controlled by Transepithelial Morphogen Gradient and Flow-Dependent Physical Cues in a Microengineered Gut-on-a-Chip. iScience 2019, 15, 391–406.
- Maurer, M.; Gresnigt, M.S.; Last, A.; Wollny, T.; Berlinghof, F.; Pospich, R.; Cseresnyes, Z.; Medyukhina, A.; Graf, K.; Gröger, M.; et al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials 2019, 220, 119396.
- De Gregorio, V.; Corrado, B.; Sbrescia, S.; Sibilio, S.; Urciuolo, F.; Netti, P.A.; Imparato, G. Intestine-on-chip device increases ECM remodeling inducing faster epithelial cell differentiation. Biotechnol. Bioeng. 2020, 117, 556–566.
- Yasotharan, S.; Pinto, S.; Sled, J.G.; Bolz, S.-S.; Günther, A. Artery-on-a-chip platform for automated, multimodal assessment of cerebral blood vessel structure and function. Lab Chip 2015, 15, 2660–2669.
- Pauty, J.; Usuba, R.; Cheng, I.G.; Hespel, L.; Takahashi, H.; Kato, K.; Kobayashi, M.; Nakajima, H.; Lee, E.; Yger, F.; et al. A Vascular Endothelial Growth Factor-Dependent Sprouting Angiogenesis Assay Based on an In Vitro Human Blood Vessel Model for the Study of Anti-Angiogenic Drugs. EBioMedicine 2018, 27, 225–236.
- Phan, D.T.T.; Wang, X.; Craver, B.M.; Sobrino, A.; Zhao, D.; Chen, J.C.; Lee, L.Y.N.; George, S.C.; Lee, A.P.; Hughes, C.C.W. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip 2017, 17, 511–520.
- Jang, K.-J.; Suh, K.-Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42.
- Vernetti, L.A.; Senutovitch, N.; Boltz, R.C.; DeBiasio, R.; Shun, T.Y.; Gough, A.; Taylor, D.L. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp. Biol. Med. 2016, 241, 101–114.
- Jiang, X.; Ren, L.; Tebon, P.; Wang, C.; Zhou, X.; Qu, M.; Zhu, J.; Ling, H.; Zhang, S.; Xue, Y.; et al. Cancer-on-a-Chip for Modeling Immune Checkpoint Inhibitor and Tumor Interactions. Small 2021, 17, 2004282.
- Bhattacharya, S.; Calar, K.; Evans, C.; Petrasko, M.; De La Puente, P. Bioengineering the Oxygen-Deprived Tumor Microenvironment Within a Three-Dimensional Platform for Studying Tumor-Immune Interactions. Front. Bioeng. Biotechnol. 2020, 8, 1040.
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Cancer 2019, 19, 65–81.
- Zhang, Y.S.; Zhang, Y.N.; Zhang, W. Cancer-on-a-chip systems at the frontier of nanomedicine. Drug Discov. Today 2017, 22, 1392–1399.
- Rodenhizer, D.; Dean, T.; D’Arcangelo, E.; McGuigan, A.P. The Current Landscape of 3D In Vitro Tumor Models: What Cancer Hallmarks Are Accessible for Drug Discovery? Adv. Healthc. Mater. 2018, 7, e1701174.
- Papapetrou, E.P. Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat. Med. 2016, 22, 1392–1401.
- Ayuso, J.M.; Virumbrales-Munoz, M.; McMinn, P.H.; Rehman, S.; Gomez, I.; Karim, M.R.; Trusttchel, R.; Wisinski, K.B.; Beebe, D.J.; Skala, M.C. Tumor-on-a-chip: A microfluidic model to study cell response to environmental gradients. Lab Chip 2019, 19, 3461–3471.
- Tsai, H.F.; Trubelja, A.; Shen, A.Q.; Bao, G. Tumour-on-a-chip: Microfluidic models of tumour morphology, growth and microenvironment. J. R. Soc. Interface 2017, 14, 20170137.
- Portillo-Lara, R.; Annabi, N. Microengineered cancer-on-a-chip platforms to study the metastatic microenvironment. Lab Chip 2016, 16, 4063–4081.
- Lee, E.; Song, H.-H.G.; Chen, C.S. Biomimetic on-a-chip platforms for studying cancer metastasis. Curr. Opin. Chem. Eng. 2016, 11, 20–27.
- Trujillo-de Santiago, G.T.; Flores-Garza, B.G.; Tavares-Negrete, J.A.; Lara-Mayorga, I.M.; González-Gamboa, I.; Zhang, Y.S.; Rojas-Martínez, A.; Ortiz-López, R.; Álvarez, M.M. The Tumor-on-Chip: Recent Advances in the Development of Microfluidic Systems to Recapitulate the Physiology of Solid Tumors. Materials 2019, 12, 2945.
- Piotrowski-Daspit, A.S.; Tien, J.; Nelson, C.M. Interstitial fluid pressure regulates collective invasion in engineered human breast tumors via Snail, vimentin, and E-cadherin. Integr. Biol. 2016, 3, 319–331.
- Sleeboom, J.J.F.; Amirabadi, H.E.; Nair, P.; Sahlgren, C.; Toonder, J.M.J.D. Metastasis in context: Modeling the tumor microenvironment with cancer-on-a-chip approaches. Dis. Model. Mech. 2018, 11, 033100.
- Avendano, A.; Cortes-Medina, M.; Song, J. Application of 3-D Microfluidic Models for Studying Mass Transport Properties of the Tumor Interstitial Matrix. Front. Bioeng. Biotechnol. 2019, 7, 6.
- Huang, C.P.; Lu, J.; Seon, H.; Lee, A.; Flanagan, L.A.; Kim, H.-Y.; Putnam, A.; Jeon, N.L. Engineering microscale cellular niches for three-dimensional multicellular co-cultures. Lab Chip 2009, 9, 1740–1748.
- Drifka, C.R.; Eliceiri, K.; Weber, S.M.; Kao, W.J. A bioengineered heterotypic stroma–cancer microenvironment model to study pancreatic ductal adenocarcinoma. Lab Chip 2013, 13, 3965–3975.
- Brancato, V.; Gioiella, F.; Imparato, G.; Guarnieri, D.; Urciuolo, F.; Netti, P. 3D breast cancer microtissue reveals the role of tumor microenvironment on the transport and efficacy of free-doxorubicin in vitro. Acta Biomater. 2018, 75, 200–212.
- Gioiella, F.; Urciuolo, F.; Imparato, G.; Brancato, V.; Netti, P.A. An Engineered Breast Cancer Model on a Chip to Replicate ECM-Activation In Vitro during Tumor Progression. Adv. Healthc. Mater. 2016, 5, 3074–3084.
- Lee, S.H.; Sung, J.H. Organ-on-a-Chip Technology for Reproducing Multiorgan Physiology. Adv. Healthc. Mater. 2017, 7, 1700419.
- Hachey, S.J.; Hughes, C.C.W. Applications of tumor chip technology. Lab Chip 2018, 18, 2893–2912.
- Kumar, V.; Varghese, S. Ex Vivo Tumor-on-a-Chip Platforms to Study Intercellular Interactions within the Tumor Microenvironment. Adv. Healthc. Mater. 2018, 8, 1801198.
- Nashimoto, Y.; Okada, R.; Hanada, S.; Arima, Y.; Nishiyama, K.; Miura, T.; Yokokawa, R. Vascularized cancer on a chip: The effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials 2019, 229, 119547.
- Maulana, T.I.; Kromidas, E.; Wallstabe, L.; Cipriano, M.; Alb, M.; Zaupa, C.; Hudecek, M.; Fogal, B.; Loskill, P. Immunocompetent cancer-on-chip models to assess immuno-oncology therapy. Adv. Drug Deliv. Rev. 2021, 173, 281–305.
- Garziano, A.; Urciuolo, F.; Imparato, G.; Martorina, F.; Corrado, B.; Netti, P. A micro-perfusion bioreactor for on line investigation of ECM remodeling under hydrodynamic and biochemical stimulation. Lab Chip 2016, 16, 855–867.
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668.
- Robertson, S.; Azizpour, H.; Smith, K.; Hartman, J. Digital image analysis in breast pathology—From image processing techniques to artificial intelligence. Transl. Res. 2018, 194, 19–35.
- Rabbani, M.; Kanevsky, J.; Kafi, K.; Chandelier, F.; Giles, F.J. Role of artificial intelligence in the care of patients with nonsmall cell lung cancer. Eur. J. Clin. Investig. 2018, 48, e12901.
- Sanghvi, A.B.; Allen, E.Z.; Callenberg, K.M.; Pantanowitz, L. Performance of an artificial intelligence algorithm for reporting urine cytopathology. Cancer Cytopathol. 2019, 127, 658–666.
- Ramón y Cajal, S.; Hümmer, S.; Peg, V.; Guiu, X.M.; De Torres, I.; Castellvi, J.; Martinez-Saez, E.; Hernandez-Losa, J. Integrating clinical, molecular, proteomic and histopathological data within the tissue context: Tissunomics. Histopathology 2019, 75, 4–19.
- Elmusrati, M.; Ashammakhi, N. Cancer-on-a-Chip and Artificial Intelligence: Tomorrow’s Cancer Management. J. Craniofacial Surg. 2018, 29, 1682–1683.
- Fetah, K.L.; DiPardo, B.J.; Kongadzem, E.M.; Tomlinson, J.S.; Elzagheid, A.; Elmusrati, M.; Khademhosseini, A.; Ashammakhi, N. Cancer Modeling-on-a-Chip with Future Artificial Intelligence Integration. Small 2019, 15, 1901985.
- Ahadian, S.; Civitarese, R.; Bannerman, D.; Mohammadi, M.H.; Lu, R.; Wang, E.; Davenport-Huyer, L.; Lai, B.; Zhang, B.; Zhao, Y.; et al. Organ-On-A-Chip Platforms: A Convergence of Advanced Materials, Cells, and Microscale Technologies. Adv. Healthc. Mater. 2018, 7, 1700506.
- De Chiara, F.; Ferret-Miñana, A.; Ramón-Azcón, J. The Synergy between Organ-on-a-Chip and Artificial Intelligence for the Study of NAFLD: From Basic Science to Clinical Research. Biomedicines 2021, 9, 248.
- Oliver, C.R.; Altemus, M.A.; Westerhof, T.M.; Cheriyan, H.; Cheng, X.; Dziubinski, M.; Wu, Z.; Yates, J.; Morikawa, A.; Heth, J.; et al. A platform for artificial intelligence based identification of the extravasation potential of cancer cells into the brain metastatic niche. Lab Chip 2019, 19, 1162–1173.
- Wang, G.; Sun, Y.; Jiang, S.; Wu, G.; Liao, W.; Chen, Y.; Lin, Z.; Liu, Z.; Zhuo, S. Machine learning-based rapid diagnosis of human borderline ovarian cancer on second-harmonic generation images. Biomed. Opt. Express 2021, 12, 5658–5669.
- Leong, T.K.M.; Lo, W.S.; Lee, W.E.Z.; Tan, B.; Lee, X.Z.; Lee, L.W.J.N.; Lee, J.-Y.J.; Suresh, N.; Loo, L.-H.; Szu, E.; et al. Leveraging advances in immunopathology and artificial intelligence to analyze in vitro tumor models in composition and space. Adv. Drug Deliv. Rev. 2021, 177, 113959.
- Pape, J.; Emberton, M.; Cheema, U. 3D Cancer Models: The Need for a Complex Stroma, Compartmentalization and Stiffness. Front. Bioeng. Biotechnol. 2021, 9, 276.
- Shukla, V.C.; Kuang, T.-R.; Senthilvelan, A.; Higuita-Castro, N.; Duarte-Sanmiguel, S.; Ghadiali, S.N.; Gallego-Perez, D. Lab-on-a-Chip Platforms for Biophysical Studies of Cancer with Single-Cell Resolution. Trends Biotechnol. 2018, 36, 549–561.
- Ingber, D.E. Is it Time for Reviewer 3 to Request Human Organ Chip Experiments Instead of Animal Validation Studies? Adv. Sci. 2020, 7, 2002030.
- Trapecar, M.; Wogram, E.; Svoboda, D.; Communal, C.; Omer, A.; Lungjangwa, T.; Sphabmixay, P.; Velazquez, J.; Schneider, K.; Wright, C.W.; et al. Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Sci. Adv. 2021, 7, eabd1707.
- Yuzhalin, A.E. Parallels between the extracellular matrix roles in developmental biology and cancer biology. Semin. Cell Dev. Biol. 2021.
More
