Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Jessie Wu and Version 1 by Hsieh Ching-Hung.
Immunotherapy is currently the backbone of new drug treatments for many cancer patients. CXC chemokine ligand 13 (CXCL13) is an important factor involved in recruiting immune cells that express CXC chemokine receptor type 5 (CXCR5) in the tumor microenvironment and serves as a key molecular determinant of tertiary lymphoid structure (TLS) formation.
- CXCL13/CXCR5
- immune cells
- cancer
1. The Expression and Implications of CXCL13/CXCR5
CXCL13 is a chemoattractant that selectively interacts with its receptor, CXCR5, to promote the migration of CXCR5+ cells toward high CXCL13 concentration areas. CXCL13 was originally identified as B cell attracting chemokine 1(BCA-1) and was shown to act on CXCR5+ B cells to induce chemotaxis and Ca2+ mobilization [81][1]. In lymph nodes, the CXCL13/CXCR5 axis is essential for homing B lymphocytes to lymphoid tissue, and CXCL13/CXCR5 deficient mice show the disrupted localization of B cells [81][1].
CXCL13 recruits B cells to tumors, where they form tertiary lymphoid structures (TLSs) [82][2]. TLSs are similar to secondary lymphoid organs on the cellular, morphological, and molecular levels, and TLSs are considered with good prognostic markers, commonly associated with better survival rates. However, the mechanisms underlying TLS development remain unclear. Rodriguez et al. found that the organization of cancer-associated fibroblasts into reticular networks depends on tumor necrosis factor receptor signaling, which acts as a lymphoid tissue organizer [83][3]. Cancer-associated fibroblasts secrete CXCL13 to recruit CXCR5+ B cells expressing lymphotoxin-α1β2, which expands TLSs in the tumor microenvironment. Moreover, CD8+ T cells mediate cancer-associated fibroblast organization, serving as an inducer of lymphoid structures to drive the formation of TLSs.
CXCR5+ T cells were originally defined as CD4+ memory-like T cells, which function to assist B cells with antibody production. The colocalization of CXCL13, CXCR5+ T cells, and B cells was detected in the follicular mantle zone. A unique subset of T cells that assist in B cell function was identified as TFH cells [84][4]. High levels of BCL6 drive TFH differentiation, and Blimp-1 is able to block T cells from differentiating into TFH cells [71][5].
TFH cells are the major CXCL13-producing cells in the lymphoid structure. In tumor microenvironments, tumor-infiltrating CXCL13-producing TFH cells may be involved in promoting local B cell localization, differentiation, and maturation [85][6]. An activated germinal center of B cells then contributes to the formation of TLSs in tumors. Gu-Trantien et al. first observed CXCL13 producing TFH cells in breast cancer and found that the TFH signature strongly predicted a positive clinical outcome in breast cancer [86][7]. Naïve CD4+ T cells differentiate into T helper 1 (Th1)-oriented TFH cells that produce CXCL13 in tumor microenvironments after being primed by antigen-presenting cells in the lymph node [87,88][8][9]. The balance of TFH/Th1 differentiation from precursor cells is determined by the competitive expression of Bcl6 and T-bet [71,89,90,91][5][10][11][12]. Further investigation revealed that tumor-infiltrating TFH cells produce CXCL13 to attract CXCR5+ CD8+ T cells and CXCR5+ B cells toward the germinal centers within the TLS, where both T cells and B cells are primed, reinforcing their cytotoxic capacity against cancer cells [92][13]. These studies indicate that BCL6-dominant signaling promotes CXCL13 expression in TFH cells [93][14]. Another subset of TFH cells, which were PD-1hiCXCR5−CD4+ T cells that expressed CXCL13, did not show elevated BCL6 expression.
In addition to TFH cells, there are several CXCL13 producing cells in lymphoid tissue and would secrete CXCL13 to chemoattract cells that express CXCR5. Denton et al. identified pulmonary fibroblasts as CXCL13-producing cells induced by Type I interferon (Type I IFN) during Influenza A virus transfection, which facilitate the recruitment of CXCR5+ B cells and T cells to the lungs to generate pulmonary germinal centers [94][15].
Antigen-specific CD4+ T cells mediate and coordinate immune cell functions in antitumor activities. Although the effects of ICI have been thoroughly explored in CD8+ T cells, the roles played by CD4+ T cells in response to ICI remain poorly understood. Balança et al. studied exhausted CD4+ T cells in head and neck, cervical, and ovarian cancers [95][16]. With exhausted CD4+ T cells defined as those cells presenting high PD-1 and CD39 levels, which were found to secrete CXCL13 and express the transcription factor thymocyte selection-associated high mobility group box (TOX). The transcription factor SRY-box transcription factor 4 (SOX-4) mediates CXCL13 production on CD4+ T cells, enriching transforming growth factor-β (TGF-β) and reducing IL-2 expression [96][17]. Similar to CXCL13+ exhausted CD8+ T cells, exhausted CD4+ T cells recruit B cells and promote the formation of TLS, suggesting a good response to immunotherapy and indicating a crucial role for CD4+ T cells in mediating the functions of various immune cells.
Cytotoxic CD8+ T cells are the strongest effectors, playing important roles in the anticancer immune response. When examining the CXCL13/CXCR5 axis, Workel et al. found that TGF-β dependent CD103+CD8+ tumor-infiltrating T cells represent an important CXCL13 resource [97][18]. When TGF-β receptors are inhibited, CXCL13 signaling is abrogated. CXCL13+CD103+CD8+ tumor-infiltrating T cells are associated with B cell recruitment, TLS formation, and neoantigen burden. Additionally, Thommen et al. also found that PD-1highCD8+ tumor-infiltrating T cells produce more CXCL13 than PD-1-CD8+ T cells in non-small-cell lung cancer (NSCLC) [34][19]. They also observed that CXCL13 recruited other CXCR5-expressing immune cells and induced the formation of TLSs in NSCLC. PD-1highCD8+ tumor-infiltrating T cells are enriched inside the TLS, and their presence successfully predicts the response to anti-PD-1 and is positively correlated with overall survival.
In chronic hepatitis B infection, Li et al. found that higher levels of CXCL13 facilitate the recruitment of CXCR5+CD8+ T cells to liver tissue in patients, which is associated with a favorable response to antiviral drug treatment [98][20]. CXCR5+CD8+ T cells secrete hepatitis B virus (HBV)-specific cytokines, such as IL-2, IFN-γ, IL-17, and IL-21. The subset of CD8+ T cells is partially exhausted but not dysfunctional and is involved in controlling the viral infection load. Strikingly, the adoptive transfer of CXCR5+CD8+ T cells to an HBV mouse model resulted in a significant decrease in HBV antigen expression. Finally, they found that B cell-deficient mice have a lower frequency of CXCR5+CD8+ T cells and decreased the HBV-specific IFN-γ+ CXCR5+CD8+ T cells in the blood and liver. These data demonstrated that B cells are required for CXCR5+ CD8+ T cells, contributing to the functions and activities of CXCR5+CD8+ T cells.
In classical Hodgkin lymphoma, Le et al. identified a special CD8+ T cell subset expressing CXCR5 and an inducible T cell costimulator (ICOS) [99][21]. This subset was functionally similar to TFH cells, with low CCR7 expression and high levels of BCL6, PD-1, CD200, and OX40 expression. In addition, these cells displayed poor cytotoxic function and low interferon-secretion and produced IL-4, IL-21, and CXCL13, similar to CD4+ TFH cells. Gene profiling analyses demonstrated that CXCR5+ICOS+CD8+ T cells are significantly similar to CD4+ TFH cells and are involved in the generation of lymphoma tumors with residual germinal centers. Overall, CD8+ T cell differentiation pathways are functionally similar to those involved in CD4+ TFH cell differentiation in Hodgkin lymphoma.
Follicular dendritic cells represent the primary cellular source of cytokines in lymphoid organs [100][22]. Previous research has identified that various dendritic cell subsets are able to secrete CXCL13, playing an important role in the establishment of interactions between lymphocytes and dendritic cells [101][23]. Additionally, CXCL13 serves as a plasma biomarker that reflects germinal center activity [102][24]. Follicular dendritic cells represent the primary source of CXCL13 expression in reactive tonsils and lymph nodes located in the B cell zone. Moreover, dysplastic and neoplastic follicular dendritic cells are able to secrete CXCL13 to recruit lymphocytes to areas of dense inflammatory infiltration [100][22]. Overall, dendritic cells secrete CXCL13 to recruit CXCR5+ lymphocytes, orchestrating the immune response in lymphoid tissues.
We summarize prior findings regarding CXCL13 and CXCR5 expression. CD8+ and CD4+ T cells, cancer-associated fibroblasts, cancer cells, and dendritic cells are able to secrete CXCL13, and CD8+ and CD4+ T cells, B cells, and cancer cells express CXCR5 (Figure 1). Increasing research has reported the detection of CXCL13 in the tumor microenvironment of many different types of cancer [103,104,105,106,107,108,109][25][26][27][28][29][30][31]. When the cancer microenvironment is enriched in CXCL13, the recruitment of CXCR5-expressing leukocytes to the tumor microenvironment increases. B cells are recruited by CXCL13 to promote TLS formation in the tumor microenvironment (Figure 2). Previous studies have correlated TLS formation with better prognosis among patients with cancer [110,111,112,113][32][33][34][35]; therefore, treatments that promote the development of TLSs represent potential therapeutic strategies.
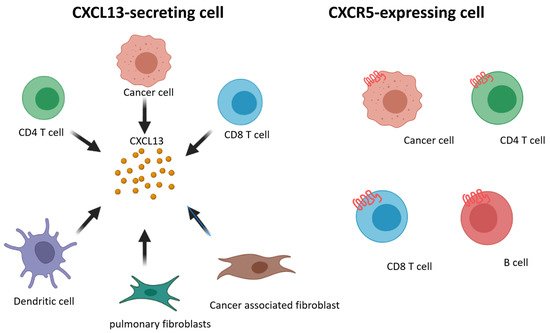
Figure 1. Schematic representation of CXCL13 and CXCR5 expressing on different cell subsets.
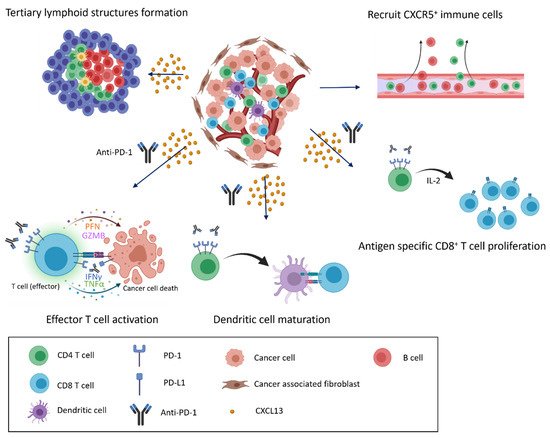
Figure 2. CXCL13/CXCR5 signaling and response to immune checkpoint blockade in the tumor microenvironment. CXCL13 secretion and enrichment in the tumor microenvironment alter the immune cell composition. CXCL13 recruits cells that express CXCR5 to infiltrate the cancer microenvironment, inducing the formation of tertiary lymphoid structures and the further infiltration of various immune cells (top). When anti-PD-1 antibodies are present, recruited immune cells actively attack cancer cells, and exhausted cells transition into effector cells, leading to the proliferation of CD8+ T cells and the maturation of dendritic cells in response to anti-PD-1 treatment (bottom).
2. CXCL13/CXCR5 Axis for ICI Response in Clinical Tumors
Immune checkpoint blockade can regulate tumor progression. Compared with chemotherapy, ICI therapy has a superior duration of response (20.4 months vs. 6.3 months) [127,128][36][37]. However, only 15–20% of patients benefit from immunotherapy [128][37] and the identification of biomarkers able to predict therapeutic response to ICI. Recently, some biomarkers were identified that were able to accurately predict the response of cancer patients to PD-1 blockade. Le et al. proposed that patients with mismatch repair deficiencies have a higher response rate to pembrolizumab treatment than patients with proficient mismatch repair processes [129][38]. In this section, we summarize clinical trials and research linking the CXCL13/CXCR5 axis with the ICI response in patients (Table 1).
Table 1. Cancer immunotherapy approaches relative to the CXCL13/CXCR5 axis based on clinical data.
| Target in the Axis | Treatment | Disease | Method of Detection | Number of Patients Investigated | Value | Outcome | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CXCL13 | + | PD1 | + | CD8 | + | T cells |
Anti-PD-1 | Non-small cell lung cancer | Transcriptome analysis | Peripheral blood of healthy donors ( | n | = 6) Fraction of PD-1 | bright | within CD8 | + | TILs ( | n | = 24) | Favorable | The presence of PD-1 | + | CD8 | + T cells can predict PD-1 blockade response and survival rate [34]. | T cells can predict PD-1 blockade response and survival rate [19]. |
| CXCL13 | Anti-PD-1 Anti-PD-L1 |
Metastatic urothelial carcinoma and bladder cancer | Whole-exome sequencing data analysis TCGA analysis |
CheckMate275 ( | n | = 270) IMvigor210 ( | n | = 310) | Favorable | CXCL13 expression plus ARID1A mutation work together to predict a favorable response to anti-PD-1 blockade [109]. | CXCL13 expression plus ARID1A mutation work together to predict a favorable response to anti-PD-1 blockade [31]. | |||||||||||||
| CXCL13 | Anti-PD-L1 | Bladder cancer | Single-sample GSEA Gene ontology analysis KEGG analysis WGCNA |
IMvigor210 ( | n | = 310) | Favorable | CXCL13 expression plus TLS formation predict a favorable response to anti-PD-1 blockade [130]. | CXCL13 expression plus TLS formation predict a favorable response to anti-PD-1 blockade [39]. | |||||||||||||||
| CXCL13 | + | /LAG3 | + | CD8 | + | T cells | Anti-PD-1 Anti-PD-L1 |
Hepatocellular carcinoma | Multiplex immunofluorescence staining TCGA-LIHC analysis Nanostring RNA analysis |
Cohort 1 ( | n | = 24) Cohort 2 ( | n | = 18) | Favorable | CXCL13 expression plus exhausted T cells marker expression predict a favorable response to anti-PD-1 blockade [125]. | CXCL13 expression plus exhausted T cells marker expression predict a favorable response to anti-PD-1 blockade [40]. | |||||||
| CXCL13 | + | CD8 | + | T cells CXCL13 | + | CD4 | + | T cells | Anti-PD-1 Nab-Paclitaxel |
Triple-negative breast cancer | ATAC-seq RNA-seq Single-cell RNA seq Whole-exome sequencing IHC |
n | = 22 | Favorable | High levels of baseline CXCL13 | + T cells predict favorable response to anti-PD-L1 plus nab-paclitaxel combination therapy [131]. | T cells predict favorable response to anti-PD-L1 plus nab-paclitaxel combination therapy [41]. | |||||||
| CXCL13 in CD8 | + | T cells | Anti-PD-L1, Anti-PD-1, Anti-CTLA4 |
Seven cancer types | Single-cell RNA-seq ATAC-seq |
n | = 1008 | Favorable | CXCL13 expression is a marker of clonal neoantigen-specific CD8 | + TILs that selectively expresses in CPI responders (“CR/PR”). [132]. | TILs that selectively expresses in CPI responders (“CR/PR”). [42]. |
|||||||||||||
| CXCL13 in tumor cells | Anti-PD-1 | Pan-cancer | Nanostring RNA analysis IHC Gene expression profiles |
NCT01295827 ( | n | = 1260) NCT01848834 ( | n | =297) NCT02054806 ( | n | = 477) | Favorable | T cells expanded signature including CXCL13 and 17 other genes are necessary for clinical response to PD-1 checkpoint blockade [133]. | T cells expanded signature including CXCL13 and 17 other genes are necessary for clinical response to PD-1 checkpoint blockade [43]. |
CXCL, CXC chemokine ligand; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; CTLA4, cytotoxic T lymphocyte-associated protein 4; RNA-seq, RNA-sequencing; ATAC-seq, Assay for Transposase-Accessible Chromatin with high-throughput sequencing; CPI, checkpoint inhibitor; HCC, hepatocellular carcinoma; TLS, tertiary lymphoid structure; ARID1A, AT-rich interactive domain-containing protein 1A; TIL, tumor-infiltrating lymphocyte; IHC, immunohistochemistry; WGCNA, Weighted correlation network analysis; TCGA, The Cancer Genome Atlas; TCGA-LIHC, The Cancer Genome Atlas Liver Hepatocellular Carcinoma; KEGG, Kyoto Encyclopedia of Genes and Genomes.
2.1. Breast Cancer
Gu-Trantien et al. analyzed CD4+ T cells in tumor tissues by profiling gene signatures. They identified TFH-related genes, such as CD200, CXCL13, ICOS, and PD-1, that were over-expressed in infiltrated tumors. They divided patients’ tumors into those with a high level of CD4+ tumor-infiltrating lymphocytes (TILs) and those with minimal infiltration [86][7]. Among patients with a high level of TILs, CXCL13 was the most highly overexpressed gene. IHC staining indicated that CXCL13 was not expressed in tumor cells but was detected intensively in TLSs and CD4+ TFH cells. Clinical correlations with CXCL13 were performed by investigating the disease-free survival (DFS) of 794 breast cancer patients, which revealed that CXCL13 expression was positively correlated with DFS in breast cancer, especially in the human epidermal growth factor 2 (HER2+) group. The authors also analyzed a cohort of 996 patients with breast cancer who received neoadjuvant chemotherapy, which showed that CXCL13 expression was highly correlated with a complete response in HER2+ patients.
Zhang et al. designed a clinical trial to identify key immune subpopulations associated with clinical outcomes for anti-PD-L1 blockade therapy in triple-negative breast cancer (TNBC) [131][41]. Twenty-two patients with late-stage TNBC were enrolled in this cohort, divided into two groups with equal numbers. Patients in one group were treated with paclitaxel, whereas the other group received paclitaxel plus atezolizumab. After 4 weeks, tumor biopsies and peripheral blood cells were collected from both groups. TILs were collected and analyzed by single-cell RNA-sequencing, single-cell ATAC-sequencing, and TCR-sequencing. In this cohort, the authors developed two indexes to associate immune subsets with clinical benefits in TNBC: the predictive index (Pi) and the therapeutic index (Ti). The Pi correlated the compositions of TIL subsets with changes in tumor volume, whereas the Ti correlated between TIL subsets dynamics with tumor volume changes. They identified B cells as being the most predictable subset for predicting the response to immune checkpoint blockade in the Pi analysis, whereas T cells were the most predictable subset to predict immune checkpoint blockade in the Ti analysis, indicating a functional change in T cells during ICI therapy. The authors clustered T cells using a high-resolution T cell map, which identified expanded CXCL13+CD8+ and CXCL13+CD4+ populations in patients who responded to paclitaxel plus atezolizumab but not patients who did not respond to treatment. Furthermore, the analysis of CXCL13+CD8+ T cells using a single-cell assay for transposase-accessible chromatin using sequencing (ATAC-seq) showed that CXCL13+CD8+ cells had more accessible chromatin regions in the IFNG, GZMK, and PDCD1 loci.
2.2. Bladder Cancer
Goswami et al. analyzed the combined data from two clinical trials, CheckMate275 and IMvigor210, to determine the correlations of the AT-rich interactive domain containing protein 1A (ARID1A) mutations and CXCL13 expression with the patient’s response to immune checkpoint blockade therapy [109][31]. CheckMate 275 is a phase 2 trial of nivolumab for the treatment of metastatic urothelial carcinoma (n = 265). IMvigor210 is a phase 2 trial of atezolizumab for the treatment of advanced or metastatic urothelial bladder cancer. They identified that an ARID1A mutation alone or CXCL13-high expression is correlated with favorable responses to ICI, and ARID1A mutation in combination with high levels of CXCL13 expression could predict the better response of patients to immune checkpoint blockade therapy than ARID1A mutation in combination with low levels of CXCL13 or high levels of CXCL13 in combination with ARID1A wild-type.
2.3. Non-Small Cell Lung Cancer
Thommen et al. classified CD8+ TILs derived from patients with NSCLC into three groups based on their PD-1 surface-level expression [34][19]. PD-1- were CD8+ TILs without detectable PD-1 expression; PD-1N were CD8+ TILs with PD-1 levels similar to those observed in healthy donors; PD-1T were CD8+ TILs PD-1 with considerably high expression levels. The reactive capacity of PD-1 TILs was confirmed by co-culturing PD-1T, PD-1N, and PD-1-, respectively, with autologous tumors isolated from eight patients. Among the eight patient-derived tumors, PD-1T TILs showed strong reactivity against the tumor cells in 6 of 8 cultures. Gene expression diversity among PD-1T, PD-1N, and PD-1- populations was identified by transcriptome analysis, which revealed that CXCL13 was one of the most upregulated genes in the PD-1T subset. The authors used bead-based immunoassays to quantify inflammatory cytokine and chemokine levels in sorted PD-1T TILs after 24h of culture, identifying that the levels of CXCL13 were the highest in the array. These data indicated that PD-1T TILs are likely to secrete CXCL13 into the tumor microenvironment. Of 21 stage IV NSCLC patients receiving anti-PD-1 therapy, 7 patients were identified as responders, whereas the other 14 patients were identified as non-responders. Responders were characterized by TIL subsets with a higher percentage of PD-1T TILs and total PD-1T TIL cell numbers. Those with PD-1T subsets > 1% of total cells were associated with longer median survival than those with PD-1T subsets < 1% of total cells following the administration of anti-PD-1 therapy. IHC and digital image analysis suggested that PD-1T TILs were localized in TLSs that formed both intratumorally and peritumorally. Intensive B cell marker staining and CD4+ TFH markers colocalized with PD-1T TILs, implying that CD8+PD-1T TILs might recruit CXCR5+ B cells and TFH cells into the tumor region and that B cells, as well as TFH cells, could reinforce the antitumor capacity of CD8+ T cells.
2.4. Hepatocellular Carcinoma
Liver cancer is one of the world’s most common cancers and the second leading cause of cancer deaths, and we also have some related studies in the past [125,134,135][40][44][45]. Hsu et al. established a platform to assess various biomarkers associated with prognostic efficacy for the immunotherapy response of patients with hepatocellular carcinoma (HCC) using Nanostring RNA analysis and multiplex immunofluorescence staining methods [125][40]. In this cohort, RNA was isolated from tumor samples before patients received anti-PD-1 or combination anti-PD-1 and anti-PD-L1 blockade therapy (n = 42). RNA was then hybridized to nCounter® probes for the 770 predefined genes featured in the PanCancer Immune Profiling Panel. They found that exhausted CD8+ T cells were enriched in responders and analyzed exhausted CD8+ T cells using transcriptome analysis. In this Immune Profiling Panel, nine genes expressed on exhausted T cells were positively correlated with a better response to anti-PD-1 monotherapy or combination anti-PD-1 and anti-PD-L1 therapy, including CXCL13 gene expression. In another study, the exploratory analysis identified distinct gene signatures associated with tumor response and resistance to anti-PD-1 monotherapy in HCC patients also showed that CXCL13 was positively correlated with a better response to anti-PD-1 therapy [136][46].
References
- Legler, D.F.; Loetscher, M.; Roos, R.S.; Clark-Lewis, I.; Baggiolini, M.; Moser, B. B Cell–attracting Chemokine 1, a Human CXC Chemokine Expressed in Lymphoid Tissues, Selectively Attracts B Lymphocytes via BLR1/CXCR5. J. Exp. Med. 1998, 187, 655–660.
- Rouanne, M.; Arpaia, N.; Marabelle, A. CXCL13 shapes tertiary lymphoid structures and promotes response to immunotherapy in bladder cancer. Eur. J. Cancer 2021, 151, 245–248.
- Rodriguez, A.B.; Peske, J.D.; Woods, A.N.; Leick, K.M.; Mauldin, I.S.; Meneveau, M.O.; Young, S.J.; Lindsay, R.S.; Melssen, M.M.; Cyranowski, S.; et al. Immune mechanisms orchestrate tertiary lymphoid structures in tumors via cancer-associated fibroblasts. Cell Rep. 2021, 36, 109422.
- Schaerli, P.; Willimann, K.; Lang, A.B.; Lipp, M.; Loetscher, P.; Moser, B. Cxc Chemokine Receptor 5 Expression Defines Follicular Homing T Cells with B Cell Helper Function. J. Exp. Med. 2000, 192, 1553–1562.
- Johnston, R.J.; Poholek, A.C.; DiToro, D.; Yusuf, I.; Eto, D.; Barnett, B.; Dent, A.L.; Craft, J.; Crotty, S. Bcl6 and Blimp-1 Are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Science 2009, 325, 1006–1010.
- Gu-Trantien, C.; Migliori, E.; Buisseret, L.; de Wind, A.; Brohée, S.; Garaud, S.; Noël, G.; Chi, V.L.D.; Lodewyckx, J.-N.; Naveaux, C.; et al. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight 2017, 2, e91487.
- Gu-Trantien, C.; Loi, S.; Garaud, S.; Equeter, C.; Libin, M.; de Wind, A.; Ravoet, M.; Le Buanec, H.; Sibille, C.; Manfouo-Foutsop, G.; et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Investig. 2013, 123, 2873–2892.
- Nakayamada, S.; Kanno, Y.; Takahashi, H.; Jankovic, D.; Lu, K.T.; Johnson, T.A.; Sun, H.-W.; Vahedi, G.; Hakim, O.; Handon, R.; et al. Early Th1 Cell Differentiation Is Marked by a Tfh Cell-like Transition. Immunity 2011, 35, 919–931.
- Lönnberg, T.; Svensson, V.; James, K.R.; Fernandez-Ruiz, D.; Sebina, I.; Montandon, R.; Soon, M.S.; Fogg, L.G.; Nair, A.S.; Liligeto, U.; et al. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci. Immunol. 2017, 2, eaal2192.
- Oestreich, K.J.; Huang, A.C.; Weinmann, A.S. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J. Exp. Med. 2011, 208, 1001–1013.
- Lüthje, K.; Kallies, A.; Shimohakamada, Y.; Belz, G.; Light, A.; Tarlinton, D.; Nutt, S. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat. Immunol. 2012, 13, 491–498.
- Pepper, M.; Pagán, A.J.; Igyártó, B.Z.; Taylor, J.J.; Jenkins, M.K. Opposing Signals from the Bcl6 Transcription Factor and the Interleukin-2 Receptor Generate T Helper 1 Central and Effector Memory Cells. Immunity 2011, 35, 583–595.
- Noël, G.; Fontsa, M.L.; Garaud, S.; de Silva, P.; de Wind, A.; Eynden, G.G.V.D.; Salgado, R.; Boisson, A.; Locy, H.; Thomas, N.; et al. Functional Th1-oriented T follicular helper cells that infiltrate human breast cancer promote effective adaptive immunity. J. Clin. Investig. 2021, 131, e139905.
- Kroenke, M.A.; Eto, D.; Locci, M.; Cho, M.; Davidson, T.; Haddad, E.K.; Crotty, S. Bcl6 and Maf Cooperate to Instruct Human Follicular Helper CD4 T Cell Differentiation. J. Immunol. 2012, 188, 3734–3744.
- Denton, A.E.; Innocentin, S.; Carr, E.J.; Bradford, B.M.; Lafouresse, F.; Mabbott, N.A.; Mörbe, U.; Ludewig, B.; Groom, J.R.; Good-Jacobson, K.L.; et al. Type I interferon induces CXCL13 to support ectopic germinal center formation. J. Exp. Med. 2019, 216, 621–637.
- Balança, C.-C.; Salvioni, A.; Scarlata, C.-M.; Michelas, M.; Martinez-Gomez, C.; Gomez-Roca, C.; Sarradin, V.; Tosolini, M.; Valle, C.; Pont, F.; et al. PD-1 blockade restores helper activity of tumor-infiltrating, exhausted PD-1hiCD39+ CD4 T cells. JCI Insight 2021, 6, e142513.
- Yoshitomi, H.; Kobayashi, S.; Miyagawa-Hayashino, A.; Okahata, A.; Doi, K.; Nishitani, K.; Murata, K.; Ito, H.; Tsuruyama, T.; Haga, H.; et al. Human Sox4 facilitates the development of CXCL13-producing helper T cells in inflammatory environments. Nat. Commun. 2018, 9, 3762.
- Workel, H.H.; Lubbers, J.M.; Arnold, R.; Prins, T.M.; van der Vlies, P.; de Lange, K.; Bosse, T.; van Gool, I.C.; Eggink, F.A.; Wouters, M.C.; et al. A Transcriptionally Distinct CXCL13+CD103+CD8+ T-cell Population Is Associated with B-cell Recruitment and Neoantigen Load in Human Cancer. Cancer Immunol. Res. 2019, 7, 784–796.
- Thommen, D.S.; Koelzer, V.; Herzig, P.; Roller, A.; Trefny, M.; Dimeloe, S.; Kiialainen, A.; Hanhart, J.; Schill, C.; Hess, C.; et al. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 2018, 24, 994–1004.
- Li, Y.; Tang, L.; Guo, L.; Chen, C.; Gu, S.; Zhou, Y.; Ye, G.; Li, X.; Wang, W.; Liao, X.; et al. CXCL13-mediated recruitment of intrahepatic CXCR5+CD8+ T cells favors viral control in chronic HBV infection. J. Hepatol. 2020, 72, 420–430.
- Le, K.S.; Amé-Thomas, P.; Tarte, K.; Gondois-Rey, F.; Granjeaud, S.; Orlanducci, F.; Foucher, E.D.; Broussais, F.; Bouabdallah, R.; Fest, T.; et al. CXCR5 and ICOS expression identifies a CD8 T-cell subset with T(FH) features in Hodgkin lymphomas. Blood Adv. 2018, 2, 1889–1900.
- Vermi, W.; Lonardi, S.; Bosisio, D.; Uguccioni, M.; Danelon, G.; Pileri, S.; Fletcher, C.; Sozzani, S.; Zorzi, F.; Arrigoni, G.; et al. Identification of CXCL13 as a new marker for follicular dendritic cell sarcoma. J. Pathol. 2008, 216, 356–364.
- Vissers, J.L.M.; Hartgers, F.C.; Lindhout, E.; Figdor, C.G.; Adema, G.J. BLC (CXCL13) is expressed by different dendritic cell subsets in vitro and in vivo. Eur. J. Immunol. 2001, 31, 1544–1549.
- Havenar-Daughton, C.; Lindqvist, M.; Heit, A.; Wu, J.E.; Reiss, S.M.; Kendric, K.; Bélanger, S.; Kasturi, S.P.; Landais, E.; Akondy, R.S.; et al. CXCL13 is a plasma biomarker of germinal center activity. Proc. Natl. Acad. Sci. USA 2016, 113, 2702–2707.
- Feng, C.; Xu, Y.; Liu, Y.; Zhu, L.; Wang, L.; Cui, X.; Lu, J.; Zhang, Y.; Zhou, L.; Chen, M.; et al. Gene Expression Subtyping Reveals Immune alterations:TCGA Database for Prognosis in Ovarian Serous Cystadenocarcinoma. Front. Mol. Biosci. 2021, 8, 619027.
- Zanetti, C.; Kumar, R.; Ender, J.; Godavarthy, P.S.; Hartmann, M.; Hey, J.; Breuer, K.; Weissenberger, E.S.; Minciacchi, V.R.; Karantanou, C.; et al. The age of the bone marrow microenvironment influences B-cell acute lymphoblastic leukemia progression via CXCR5-CXCL13. Blood 2021, 138, 1870–1884.
- Zhou, X.; Peng, M.; He, Y.; Peng, J.; Zhang, X.; Wang, C.; Xia, X.; Song, W. CXC Chemokines as Therapeutic Targets and Prognostic Biomarkers in Skin Cutaneous Melanoma Microenvironment. Front. Oncol. 2021, 11, 619003.
- Lv, Y.; Lv, D.; Lv, X.; Xing, P.; Zhang, J.; Zhang, Y. Immune Cell Infiltration-Based Characterization of Triple-Negative Breast Cancer Predicts Prognosis and Chemotherapy Response Markers. Front. Genet. 2021, 12, 616469.
- Li, N.; Li, B.; Zhan, X. Comprehensive Analysis of Tumor Microenvironment Identified Prognostic Immune-Related Gene Signature in Ovarian Cancer. Front. Genet. 2021, 12, 616073.
- Li, Y.; Wu, T.; Gong, S.; Zhou, H.; Yu, L.; Liang, M.; Shi, R.; Wu, Z.; Zhang, J.; Li, S. Analysis of the Prognosis and Therapeutic Value of the CXC Chemokine Family in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2021, 10, 570736.
- Goswami, S.; Chen, Y.; Anandhan, S.; Szabo, P.M.; Basu, S.; Blando, J.M.; Liu, W.; Zhang, J.; Natarajan, S.M.; Xiong, L.; et al. ARID1A mutation plus CXCL13 expression act as combinatorial biomarkers to predict responses to immune checkpoint therapy in mUCC. Sci. Transl. Med. 2020, 12, eabc4220.
- Qin, M.; Hamanishi, J.; Ukita, M.; Yamanoi, K.; Takamatsu, S.; Abiko, K.; Murakami, R.; Miyamoto, T.; Suzuki, H.; Ueda, A.; et al. Tertiary lymphoid structures are associated with favorable survival outcomes in patients with endometrial cancer. Cancer Immunol. Immunother. 2021, 1–12.
- Dieu-Nosjean, M.-C. Tumor-Associated Tertiary Lymphoid Structures: A Cancer Biomarker and a Target for Next-generation Immunotherapy. Adv. Exp. Med. Biol. 2021, 1329, 51–68.
- Delvecchio, F.R.; Fincham, R.E.; Spear, S.; Clear, A.; Roy-Luzarraga, M.; Balkwill, F.R.; Gribben, J.G.; Bombardieri, M.; Hodivala-Dilke, K.; Capasso, M.; et al. Pancreatic Cancer Chemotherapy Is Potentiated by Induction of Tertiary Lymphoid Structures in Mice. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1543–1565.
- Wennhold, K.; Thelen, M.; Lehmann, J.; Schran, S.; Preugszat, E.; Garcia-Marquez, M.; Lechner, A.; Shimabukuro-Vornhagen, A.; Ercanoglu, M.S.; Klein, F.; et al. CD86+ Antigen-Presenting B Cells Are Increased in Cancer, Localize in Tertiary Lymphoid Structures, and Induce Specific T-cell Responses. Cancer Immunol. Res. 2021, 9, 1098–1108.
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833.
- Friedlaender, A.; Kim, C.; Addeo, A. Rethinking the Optimal Duration of Immune Checkpoint Inhibitors in Non-small Cell Lung Cancer Throughout the COVID-19 Pandemic. Front. Oncol. 2020, 10, 862.
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520.
- Chen, X.; Xu, R.; He, D.; Zhang, Y.; Chen, H.; Zhu, Y.; Cheng, Y.; Liu, R.; Zhu, R.; Gong, L.; et al. CD8+ T effector and immune checkpoint signatures predict prognosis and responsiveness to immunotherapy in bladder cancer. Oncogene 2021, 40, 6223–6234.
- Hsu, C.-L.; Ou, D.-L.; Bai, L.-Y.; Chen, C.-W.; Lin, L.; Huang, S.-F.; Cheng, A.-L.; Jeng, Y.-M.; Hsu, C. Exploring Markers of Exhausted CD8 T Cells to Predict Response to Immune Checkpoint Inhibitor Therapy for Hepatocellular Carcinoma. Liver Cancer 2021, 10, 1–14.
- Zhang, Y.; Chen, H.; Mo, H.; Hu, X.; Gao, R.; Zhao, Y.; Liu, B.; Niu, L.; Sun, X.; Yu, X.; et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 2021, 39, 1578–1593.e8.
- Litchfield, K.; Reading, J.L.; Puttick, C.; Thakkar, K.; Abbosh, C.; Bentham, R.; Watkins, T.B.K.; Rosenthal, R.; Biswas, D.; Rowan, A.; et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 2021, 184, 596–614.e14.
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940.
- Hsu, C.; Lin, L.-I.; Cheng, Y.-C.; Feng, Z.-R.; Shao, Y.-Y.; Cheng, A.-L.; Ou, D.-L. Cyclin E1 Inhibition can Overcome Sorafenib Resistance in Hepatocellular Carcinoma Cells Through Mcl-1 Suppression. Clin. Cancer Res. 2016, 22, 2555–2564.
- Lu, J.-W.; Ho, Y.-J.; Yang, Y.-J.; Liao, H.-A.; Ciou, S.-C.; Lin, L.-I.; Ou, D.-L. Zebrafish as a disease model for studying human hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 12042–12058.
- Hsu, C.; Edeline, J.; Masi, G.; Ma, Y.T.; Wang, W.; Wege, H.; Fei, C.; Ling, C.; Ma, X.; Zhang, P.; et al. 360 Tumor-immune signatures associated with response or resistance to tislelizumab in patients with previously treated advanced hepatocellular carcinoma (HCC). J. Immunother. Cancer 2021, 9, A387.
More
