Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Friedrich Altmann and Version 2 by Rita Xu.
In the animal kingdom, a stunning variety of N-glycan structures have emerged with phylogenetic specificities of various kinds. In the plant kingdom, however, N-glycosylation appears to be strictly conservative and uniform. From mosses to all kinds of gymno- and angiosperms, land plants mainly express structures with the common pentasaccharide core substituted with xylose, core α1,3-fucose, maybe terminal GlcNAc residues and Lewis A determinants. In contrast, green algae biosynthesise unique and unusual N-glycan structures with uncommon monosaccharides, a plethora of different structures and various kinds of O-methylation.
- moss
- bryophytes
- glycoprotein
- N-glycan
- methyl-mannose
1. Introduction
The many branches of the tree of life distinguish themselves—inter alia—by their characteristic sets of structural features of N-glycans. Chordates have evolved sialic acids, which are missing in all other parts of the eukaryotic world as far as authorswe currently know [1][2][1,2]. Subtle changes in the general N-glycome of vertebrates have occurred in the course of the development of vertebrates; thus, the structural repertoires of fish, birds, mammals and humans differ. Protostomia such as worms, molluscs, or arthropods impress with a variety of yet other structural peculiarities, such as phosphoethanolamine or substituted fucose residues, just to name a few examples [1]. Plants, however, appear to be highly conservative, displaying the very same set of N-glycan structures throughout the division of land plants, as was revealed by survey studies on the N-glycomes of pollen and food allergens [3][4][3,4]. The plant N-glycome was deciphered in two waves. The first one culminated in the late 1970s with the elucidation of the exact structure of the N-glycan of the pineapple stem protease bromelain [5][6][5,6]. This pioneering work was soon followed by the elucidation of the structure of the horseradish peroxidase N-glycan [7] and examples of terminal N-acetylglucosamine residues [3][8][9][10][3,8,9,10]. Here, it could be included that the xylose and even more the core α1,3-fucose residue are recognised as immunogenic and even allergy-relevant determinants [7][11][12][13][7,11,12,13].
A second wave of discovery revealed decoration with Lewis A determinants on one or two antennae [14][15][14,15]. This actually widely distributed feature [3][8][10][14][3,8,10,14] was overlooked for some time as it can hardly ever be found on high-abundance storage proteins but rather on secretory or cell wall glycoproteins [14]. The ultimate conservation of these traits points to an essential physiological function of complex-type plant glycans, which, however, have not been fathomed so far as the deletion of either xylosyl-transferase, core-α1,3-fucosyltransferase, or the Lewis A fucosyltransferase did not result in obvious phenotypic alterations [16][17][18][19][16,17,18,19]. Even a GlcNAc-transferase I-deficient Arabidopsis line, at first glance, appeared as a normal viable plant [20][21][20,21], but for tomatoes, complex glycans do play a vital role [22].
The structural investigation of the biotechnologically relevant moss species Physcomitrella patens (now Physcomitrium patens) [23] by HPLC or (low-resolution) MALDI-TOF MS displayed a picture exactly matching that seen with apple, onion, cauliflower, or any other fresh plant tissue [10]. It was the incidental appearance of an impressive amount of sporangia on a lawn of moss outside the author’s living room that enticed us to record yet another boring plant N-glycome. However, this moss—Bryum caespiticium—looked different. Now viewed with high-resolution MALDI-TOF MS, the small complex-type glycans MMXF and MGnXF (or, more precisely, MMXF3 and MGnXF3) were accompanied by prominent peaks each larger by 14.02 mass units, strongly indicative of methylation (Figure 1).
2. Discovery of Methylation of Moss N-glycans
The MALDI-TOF MS spectrum of the released N-glycans from sporangia as well as gametophytes of Bryum caespiticium exhibited eye-catching extra peaks next to the usual complex-type plant N-glycans (Figure 1A). The glycosylation pattern of sporangia and lawn did not show a significant difference (data not shown).
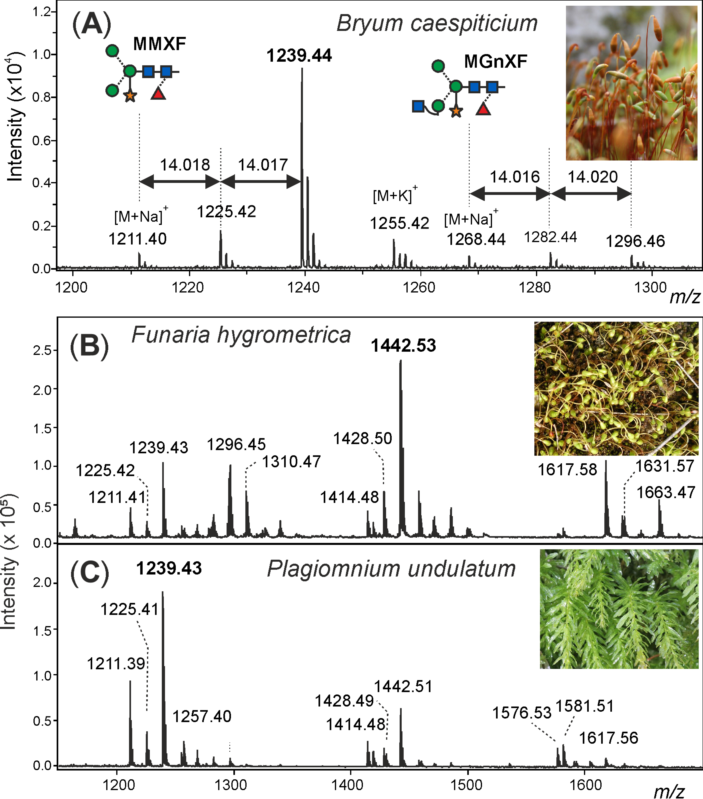
Figure 1. Demonstration of methylation of moss N-glycans. The structure abbreviations MMXF and MGnXF specify the terminal residues in a counter-clockwise manner. (A) Detail of the MALDI-TOF MS spectrum of N-glycans from the sporangia of Bryum caespiticium showing 14.02 Da increments. Section of the MALDI-TOF spectra of N-glycans from Funaria hygrometrica (B) and Plagiomnium undulatum (C) showing the different dominant complex-type N-glycans and their methylation. Complete spectra of all mosses analysed can be found in the Supporting Information.
The exact mass increment of 14.02 pointed at a methylation of the glycans, rather than another modification. After the first sign that methylated N-glycans may also exist in land plants, the question appeared, if this moss was one of its kind or if there are methylated N-glycans in other moss species as well.
3. N-glycosylation Pattern of Different Moss Species
After purifying the N-glycans from several moss species, the glycan patterns were analysed with MALDI-TOF. The main masses found represented the glycans MMXF, MGnXF and GnGnXF as well as singly and doubly methylated versions of these (Figure 1 and Figure 2). The main glycan type in many analysed moss species represented the mass of MMXF, while two of the analysed species had a main glycan that was doubly methylated (Figure 2). The moss Funaria hygrometrica presented a doubly methylated MGnXF (1442.53 Da [M+Na]+) as the main glycan (Figure 1B), whereas doubly methylated MMXF (1239.43 Da [M+Na]+) dominated in Plagiomnium undulatum (Figure 1C). In many mosses with methylated N-glycans, singly methylated MMXF and MGnXF were more abundant than the doubly methylated glycans. However, the mosses with higher methylation levels, such as Funaria hygrometrica, Tortula muralis, Ceratodon purpureus, Plagiomnium undulatum and Bryum caespiticium, tended to have a higher abundance of doubly methylated glycans compared to singly methylated ones. Hypnum cupressiforme, Vesicularia montagnei and Calliergonella cuspidata, all belonging to the order of Hypnales, had a higher abundance of singly as compared to doubly methylated glycans. For the glycan GnGnXF, only singly methylated glycans were found in all moss species with methylation. Moreover, GnGnXF consistently carried the lowest amount of methylation. The degree of methylation varied immensely between different moss species, even in mosses belonging to the same family. For this, examples are the mosses Aloina aloides and Tortula muralis, both belonging to the family of Pottiaceae. While Tortula muralis had a high amount of methylated glycans, Aloina aloides showed no methylation at all. Moreover, in the family of Funariaceae, the methylation level was heterogeneous—e.g., MGnXF was highly methylated only in Funaria hygrometrica. Physcomitrium sphaericum and Physcomitrium eurystomum also had methylated N-glycans, but the amount was low (Figure 2). Of all moss species analysed, the third Physcomitrium species, P. patens, was striking for its predominant (unmethylated) complex-type GnGnXF glycosylation. How far the methylation patterns are stable during the life cycle of the mosses or reflect different stages is currently unknown.
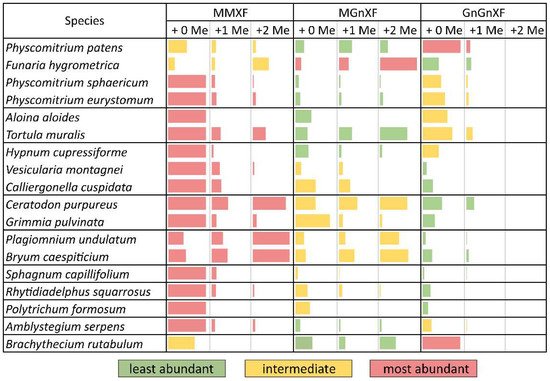

Figure 2. Glycan abundance in different moss species. Mosses from the same family are clustered together in separate boundaries. The abundance of MMXF, MGnXF and GnGnXF with their methylation is shown with coloured bars. The size of the bars is relative to the most abundant glycan from each species. Red indicates the most abundant glycan form. Physcomitrium patens was formerly known as Physcomitrella patens.
To obtain more insight into the relations between the analysed mosses, a phylogenetic tree was built with an online tool. The resulting tree nicely matched a recently published taxonomy [24][27]. Although the phylogenetic tree showed clustering, these clusters did not match the methylation status. The only cluster that showed homogeneous methylation was the cluster with Plagiomnium undulatum and Bryum caespiticium, both belonging to the order of Bryales. The other clusters were very heterogeneous in their level of methylation, which is comparable to Figure 2 (Figure 3). The samples derived from mosses adapted to both arid and highly humid habitats. They comprised gametophytes and, in case of Bryum, also sporophyte tissue. Insights into the regulation and possible biological function of N-glycan methylation have to await more systematic, ideally prospective studies.
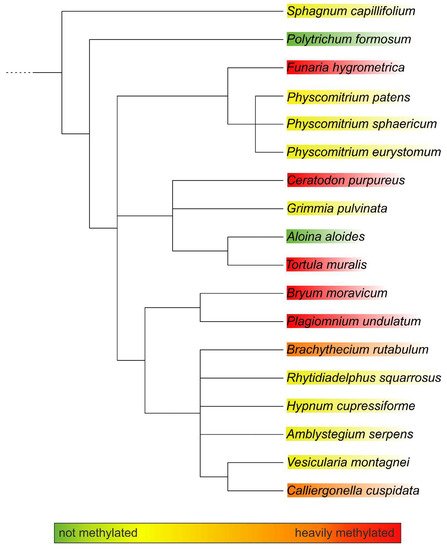

Figure 3. Phylogenetic tree of the analysed moss species as generated with PhyloT. The degree of methylation is indicated by the colour code.
4. Structural Analysis of the Main Glycans from Funaria hygrometrica and Plagiomnium undulatum
For structural analysis, the glycomes of F. hygrometrica and P. undulatum were fractionated with HILIC-HPLC to purify the doubly methylated MGnXF from F. hygrometrica and the doubly methylated MMXF from P. undulatum. The fractions containing mainly the glycans of interest were then hydrolysed, reduced and peracetylated before loading them on a GC-MS. For F. hygrometrica one part of the sample was also permethylated with iodomethane-d3, to get insight into the linkages of the monosaccharides and to confirm the overall structure of what was assumed to be the typical plant glycan MGnXF. The measurement showed that the glycan indeed adopted the well-known plant-type MGnXF structure with a doubly methylated, terminal mannose. The MS fragments located one methyl group to the C2, but gave an unclear result for the second one. A clearer identification was obtained for both the major N-glycans of F. hygrometrica and P. undulatum by the omission of the permethylation step. Fragmentation pattern and retention time clearly identified a 2,6-di-O-methyl mannose. To contest the assumption that this methyl mannose represented the 6-branch, 2me-MMXF and 2me-MGnXF were subjected to negative mode collision-induced decay in an LC-ESI-MS analysis. D-ions of m/z 351 in both glycans established the 6-arm as carrying the methyl-mannose [25][28].
The structure of the F. hygrometrica glycan is therefore MGnXF with a doubly methylated mannose on the upper arm. The structure of the main glycan from P. undulatum is an MMXF with a doubly methylated mannose on the upper arm (Figure 4). The arm location of methyl mannose was substantiated by the D-ion observed in negative mode collision-induced dissociation (Figure 5).
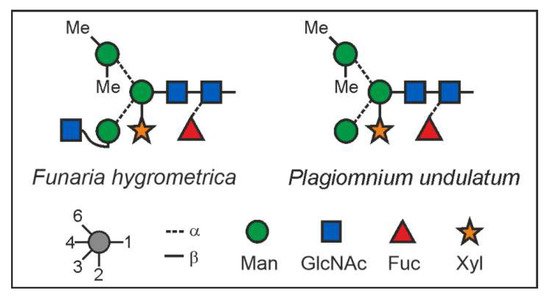
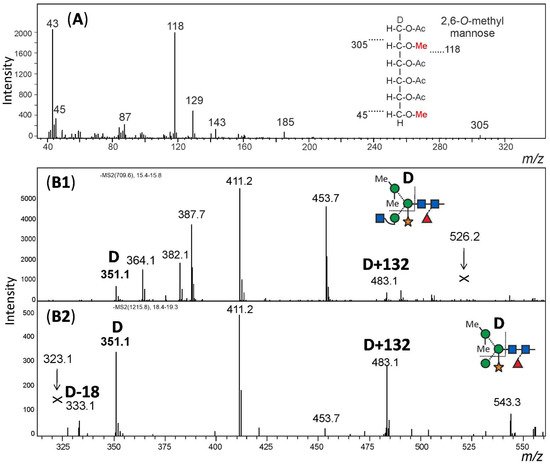

Figure 4. N-glycan structures of the main glycans from Funaria hygrometrica and Plagiomnium undulatum.

Figure 5. Structural analysis of methylated N-glycans. Panel (A): The HILIC fractionated main glycan from Funaria hygrometrica was hydrolysed, reduced and peracetylated. The fragment peaks in GC-MS analysis identified the methyl mannose as 2,6-di-O-methylation, which fits the retention time of that peak. Panels (B1,B2): The arm location of the methylated mannose was defined by observing the D-ion generated by negative mode collision induced dissociation. Notably, the xylose substituting the β-mannose residue was partially retained on the D-ions. Arrows pointing at Andrew’s crosses denote the theoretical positions of D-ion for the alternative branch arrangement.
