Toddalia is a monotypic genus consisting of sole species Toddalia asiatica (L.) Lam., widely distributed in Africa and Asia. It belongs to tribe Amyridoideae, family Rutaceae and order Sapindales. This genus together with genera Phellodendron Rupr., Tetradium Lour., and Zanthoxylum L. were found to produce alkaloids. The relationship of these chemical constituents has been hypothesized for the past years. The ancestor for Rutaceae has been linked to the several alkaloids recognized in these four genera, thus naming the group to date proto-Rutaceae. The four genera have attracted the attention of many scientists and they have been studied widely in previous studies. However, there is insufficient genomic resources in this genus. Additionally the origin and dispersal points of Toddalia species remains unclear. Here, rwesearchers sequenced the two samples of Toddalia asiatica species, both collected from Kenya, and made comparison of their genome structures with T. asiatica species from China, available in the NCBI database. Phylogenomic analysis and impacts of climate change on Toddalia species were also conducted. The availability of the sequenced cp genomes will provide valuable genetic resources for further population genetics and biogeographic studies of these species by sampling them from a wider geographical range.
1. Introduction
The family Rutaceae contains flowering plants belonging to the order Sapindales. It comprises about 150–165 genera and approximately 2100 species widely distributed in the tropical and subtropical regions.
Zanthoxylum is the second largest genus of Rutaceae, consisting of approximately 225 species, and it has a worldwide distribution
[1][2][7,8].
Rutaceae has traditionally been divided into (two, three, four, or seven) subfamilies
[3][4][5][6][3,6,9,10]. Amyridoideae is the largest and diverse subfamily based on the current circumscription
[6][10]. In this subfamily, four genera
Phellodendron Rupr.,
Tetradium Lour.,
Toddalia Juss., and
Zanthoxylum L. were found to produce alkaloids
[2][7][8][8,11,12] and the relationship of these chemical constituents has been hypothesized for the past years. The ancestor for Rutaceae has been linked to the several alkaloids recognized in these four genera, thus naming the group to date proto-Rutaceae
[9][13].
Toddalia is a monotypic genus with one species
T. asiatica (L.) Lam., widely distributed in Africa and Asia.
The four groups have been widely studied in previous studies
[3][10][3,14]. The results indicated that
Zanthoxylum formed a sister group with
Toddalia. However, it was evidenced that the genus
Toddalia was nested within
Zanthoxylum using the highest sampled taxa of
Zanthoxylum [11][15]. Additionally, studies have proposed the change of
T. asiatica to
Zanthoxylum asiaticum (L.) Appelhans, Groppo & J. Wen, comb. Nov
[2][8]. Moreover, the current research conducted using 36 complete plastomes supported
Toddalia as part of
Zanthoxylum and suggested that they should be merged
[4][6].
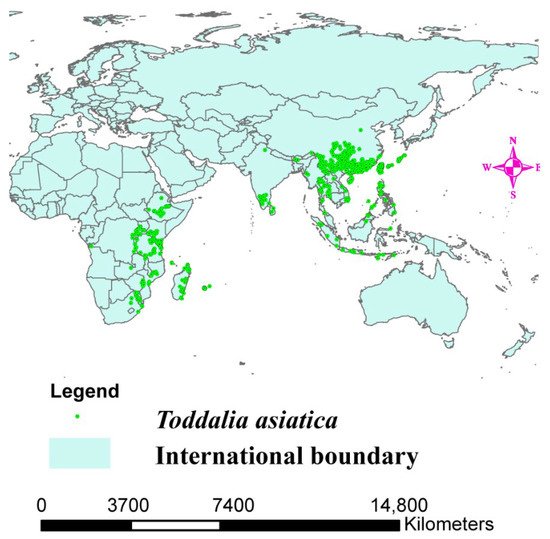
The range of Toddalia asiatica may directly change because of altered climate, resulting in a change in its distribution (Figure 1). Projecting the favorable climatic niche for these species and investigating their control range is important for estimating the effects of climate change on the species as well as knowing the conservation measures to put in place. The objectives of this study were to (1) sequence the two samples of T. asiatica species collected from Kenya, and compare their structural features with T. asiatica from Asia available in the NCBI database, (2) determine the phylogenetic placement of T. asiatica samples from Africa in the phylogenetic tree, (3) determine the origin of the genus Toddalia and its dispersal areas, and (4) determine the impacts of climate change on Toddalia species.
Figure 1. Distribution map of Toddalia asiatica in Africa and Asia.
2. Complete Chloroplast Genome
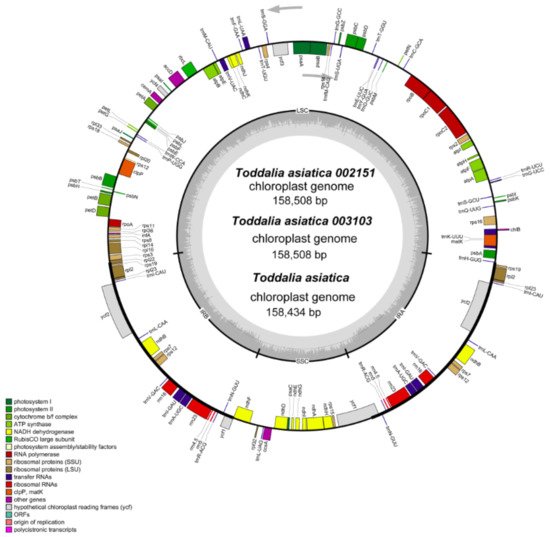
Using the advanced sequencing technology, the total DNA of the two samples of Toddalia species was isolated. The number of annotated genes for these samples is shown (Table 1). They had a total of 113 genes including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The total length of the cp genomes (T. asiatica 002151 and T. asiatica 003103), GC content, number of genes, and other information are shown (Table 2). The cp genome size for the sequenced samples was 158, 508 bp (Figure 2), which was slightly larger compared to the published genome of T. asiatica (158,434) collected from China. The cp genomes displayed a typical quadripartite structure consisting of a pair of IRs (27,007) separated by one LSC (86,162) and one SSC (18,332) region.
Figure 2. Gene map of
Toddalia plastomes. Genes in the circle are transcribed clockwise, while the rest are transcribed counterclockwise. Dark gray shading in the inner circle indicates the GC content.
Table 1. List of annotated genes of T. asiatica.
| Gene Group |
Gene Name |
| Features |
T. asiatica 002151 |
T. asiatica 003103 |
T. asiatica |
| Ribosomal RNAs |
rrn16(2), rrn23(2), rrn5(2), rrn4.5(2) |
| Transfer RNAs |
trnA-UGC*(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnH-GUG, trnS-CGA, trnK-UUU*, trnL-CAA(2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-GUA*, trnW-CCA, trnY-GUA |
| Proteins of small ribosomal subunit |
rps16*, rps2, rps14, rps15, rps4, rps7(2), rps18, rps12* (2), rps11, rps8, rps |
27,007 |
| 36, | rpl14, rpl16*, rpl22, rpl2* (2), rpl23(2), rpl32 |
| Subunits of RNA polymerase |
rpoC2, rpoC1* |
| Photosystem I |
| Total cp genome size (bp) |
158,508 |
158,508 |
158,434 |
| Length of LSC (bp) |
86,162 |
| 3, | rps19(2) |
| 27,008 |
Proteins of large ribosomal subunit |
rpl33, rpl20, |
| Length of SSC (bp) | rpl |
18,332 |
18,332 |
18,286 |
| Total GC content (%) |
38.5 |
38.5 |
38.5 |
psaB, psaA, psaI, psaJ, psaC |
| 86,162 |
| GC content of LSC (%) |
36.8 |
36.8 |
Photosystem II |
psbA, psbB, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ, psbC |
| 36.8 |
Cytochrome b/f complex |
petA, petB*, petD*, petG, petL, petN |
| Subunits of ATP synthase Protease |
atpA, atpB, atpE, atpF*, atpH, atpI clpP** |
| 86,132 |
The large subunit of rubisco |
| GC content of IR (%) |
42.9 |
42.9 |
42.9 |
| GC content of SSC (%) |
33.4 |
33.4 |
33.4 |
| Total number of genes |
113 |
113 |
115 |
rbcL |
| Protein encoding genes |
79 |
79 |
81 |
NADH dehydrogenase |
ndhA*, ndhB*(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Maturase |
matK |
| Envelope membrane protein |
cemA |
| Acetyl-CoA carboxylase |
accD |
| Synthesis gene |
ccsA |
| Open reading frames (ORF, ycf) |
ycf1, ycf2(2), ycf3**, ycf4, ycf15(2), |
Table 2. General features of the Toddalia chloroplast genomes compared in this study.
| Length of IR (bp) |
| 27,007 |
| tRNA genes |
| 30 |
| 30 |
| 30 |
| rRNA genes |
4 |
4 |
4 |
The DNA gene content (GC) is an important factor that implies the closeness of species. The GC content of
Toddalia species compared was 38.5%, which was lower than the AT content. The average GC contents of the LSC region, IR region, and SSC regions were 36.8%, 42.9%, and 33.4%. This indicates that the rRNA and tRNA, which occupy mainly IR regions, prefer to use bases G and C. However, the IR regions depicted a higher GC content of 42.9% and this is attributed to the presence of the rRNA and tRNA genes within the inverted repeat region, which occupies a greater area than the protein-coding genes. This occurrence has also been displayed in other studies
[12][24].
3. Codon Preference Analysis
Codon usage is an important aspect of gene expression in the species genomes
[13][25]. Different species tend to use some codons often while other codons are rarely used. This indicates that different organisms vary in their synonymous codon rates of occurrences in their protein-coding sequences. Genes in the closely related species generally show a similar codon use pattern. Through the analysis of codon preference,
researchwe
rs can better understand the evolution of species
[14][26]. Based on the protein-coding sequences, the frequency of codon usage was estimated for
Toddalia plastomes. In total, the protein-coding genes were composed of 52,811 for
T. asiatica and 52,836 for both
T. asiatica 002151 and
T. asiatica 003103, which encoded 21 amino acids (
Supplementary Table S2, could be found in https://www.mdpi.com/2223-7747/11/2/231#supplementary). Among them, the most frequent amino acid was leucine 10.24% for
T. asiatica 002151, 10.20% for
T. asiatica 003103, and 10.01% for
T. asiatica. The least frequent coded amino acid was cysteine with 1.385% for
T. asiatica 002151, 1.313% for
T. asiatica 003101, and 1.351% for
T. asiatica. In our analysis, the relative synonymous codon usage (RSCU) values of 35 codons were greater than 1 and most of them ended with A or U and only five codons ended with G. This indicated that the preferred codons tended to be A/U ending. This phenomenon was the same as that reported for other Rutaceae plastomes
[15][27]. The choice of codon usage is a result of mutation and selection factors
[16][28]. Thus, the choice of codons within the plastome can be used to show gene expressions and speciation mechanisms in species.
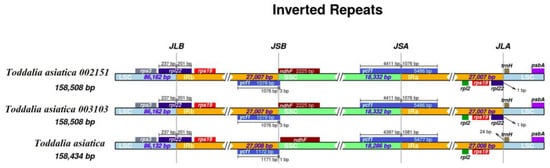
4. IR Contraction and Expansion
The contraction and expansion of the IR region at the borders affect the size difference between chloroplast genomes and play important roles in evolution
[17][29]. Although the IR regions of these species were relatively conserved, some structural variations were noted in
T. asiatica 002151 and
T. asiatica 003103 from Africa compared to the reference species from Asia (
Figure 3). The IR regions for the plastomes were 27,008 bp in length for
T. asiatica and 27,007 bp in length for
T. asiatica 002151 and
T. asiatica 003103. In these species, genes
rpl22 and
ycf1 extended in the LSC/IRb and SSC/IRa borders. Gene’s
rps3,
rps19,
rpl2,
rpl22,
trnH, and
psbA were found in the LSC/IRb and IRa/LSC region while
ndhF was found in the IRb/SSC region. In the two samples of
Toddalia species from Africa, the gene
rpl22 was found in the LSC/IRb and IRa/LSC regions. All the plastomes had similar SSC/IRa borders and the junction was all crossed by the gene ycf1 with a length from 1076 to 1081 bp. In addition, it was found that the gene
rpl2 was found in all the species. Inverted repeats are the most conserved regions of the chloroplast genome
[18][30] and great variations occur in land plants
[19][31]. They play a vital role in the stabilization of the plastome
[20][32] and also affect their size
[21][33].
Figure 3. Comparison of the borders of large single copy (LSC), small single copy, (SSC), and inverted repeat (IR) regions among the
Toddalia plastomes. Different color boxes indicate specific genes.
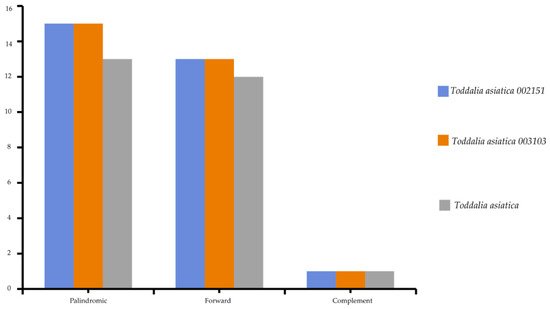
5. Repeat Analysis
Chloroplast repeats are major genetic resources that take a vital role in the rearrangement and recombination of the genomes
[22][34]. They are useful for carrying out biogeographical and population genetic studies
[23][35]. The plastomes of the species used in this study were found to contain a varied number of repeats (i.e., palindromic, forward, and reverse repeats). 81 repeats was recorded comprising of 43 palindromic repeats, 38 forward repeats, and 3 complementary repeats (
Figure 4). The long repeats in these
Toddalia species ranged from 30 to 73 bp in length. The long repeat length of 30 bp was dominant and occurred in all the species’ cp genomes (
Supplementary Table S3). The number of tandem repeats was 25 in
T. asiatica, comprising of 13 palindromic repeats, 12 forward repeats, and a complement repeat. A total of 28 repeats in both
T. asiatica 002151 and
T. asiatica 003103 consisting of 15 palindromic repeats, 13 forward repeats, and a complement repeat were recorded.
Figure 4. Total number of repeats found in
Toddalia species.
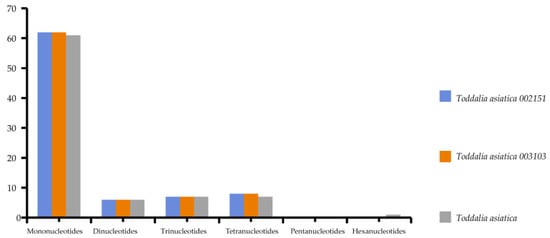
Simple sequence repeats (SSRs) are short repeat motifs of DNA sequences that normally show high levels of polymorphism, even between closely related species. Chloroplast SSRs are potentially useful molecular markers for population genetics and polymorphism studies
[24][25][26][27][28][29][36,37,38,39,40,41]. There were no great variations found in the number of SSRs calculated in these species. Mononucleotides to tetranucleotides were present in all the samples. Pentanucleotides were missing in all the species while there was only one hexanucleotide in
T. asiatica. Among these SSRs, mononucleotides reported the highest repeat motifs (
Supplementary Table S4).
Toddalia asiatica from Asia had 82 SSRs comprising of 61 mononucleotides, 6 dinucleotides, 7 trinucleotides and tetranucleotides, and 1 hexanucleotide. Both
T. asiatica 002151 and
T. asiatica 003103 from Africa had 83 SSRs with 62 mononucleotides, 6 dinucleotides, 7 trinucleotides, and 8 tetranucleotides (
Figure 5,
Supplementary Table S5). Although the genes within the chloroplast plastomes are highly conserved, the number of SSRs differs among and within the species
[29][41]. Among these samples of cp plastomes, A/T mononucleotide repeats were the highest and these findings were in concordance with other studies
[30][31][42,43]. However, other studies have indicated di-nucleotides and tri-nucleotides as the highest
[23][32][35,44]. Our findings demonstrated that SSRs within these chloroplast plastomes majorly consist of A/T repeats. This is attributed to the A/T abundance of the species cp plastomes. This phenomenon has been observed in many previous studies
[33][34][45,46]. Therefore, SSRs are important for understanding intrageneric and intergeneric differences within species.
Figure 5. The total number of simple sequence repeats present in
Toddalia species.
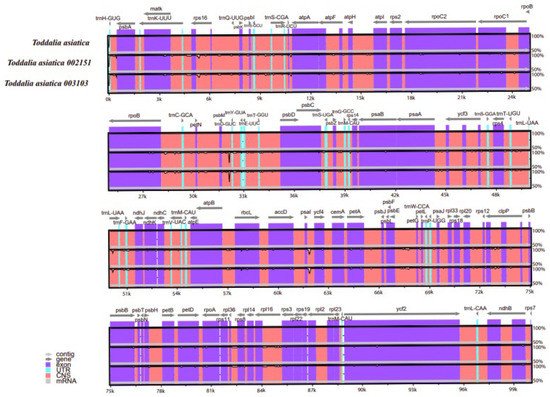
6. Comparative Analysis
Comparison of the
Toddalia samples was conducted using Mvista and with the annotation of
T. asiatica as a reference. The comparison demonstrated that the plastomes had a high consistency in gene arrangement (
Figure 6,
Supplementary Figure S1). The results depicted that the gene number, orientation, and order were highly conserved. Most sequence variations occurred in the non-coding sequences, which indicated that the coding regions were more conserved than non-coding regions. The IR regions were much less divergent than the LSC and SSC regions. Similarly, non-coding regions had higher gene differences compared to the coding regions. Moreover, within the SSC and the LSC regions, the non-coding regions were more varied than the coding regions. This pattern has been observed in other studies
[35][47]. The IR regions were more conserved based on the abundance and gene order. The differences in the size of these cp genomes could be a result of the contraction and expansion of the IR regions. DNA barcodes generally have a high mutation rate and can be utilized in the identification of species in a given taxonomic group
[36][37][38][48,49,50] and they can be vital markers for evolutionary studies as well as barcoding.
Figure 6. Sequence alignment of plastomes of
Toddalia species using the LAGAN method. A cut-off of 70% similarity was used for the plot and the y-scale represents the percent similarity ranging from 50–100%.
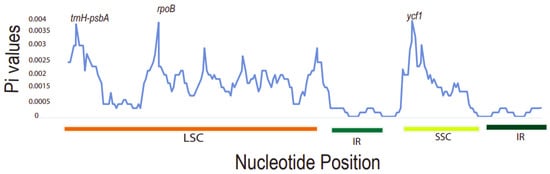
7. Sequence Divergence Analysis
It was found that the Pi values ranged from 0 to 0.004. The three most divergent hotspot regions observed were
trnH-psbA,
rpoB, and
ycf1 genes. They exhibited significantly higher Pi values (>0.0035) and were all located in the LSC and SSC region. Inverted repeat regions displayed lower nucleotide diversity, depicting that they are more conserved compared to the LSC and SSC regions (
Figure 7). The large single-copy region (LSC) had the highest number of diverse regions followed by the SSC region. The dN/dS value can be used to measure the evolution rate of a specific gene
[39][51]. This is the ratio of the synonymous substitution rate (dS) to the non-synonymous substitution rate (dN), which is important in analyzing the evolutionary pressures within the plastomes. Synonymous substitutions are more frequent than non-synonymous ones in protein-coding sequences
[40][52]. These varied regions may be undergoing rapid nucleotide substitution at the species level. Thus, this demonstrates their ability as potential barcode markers for phylogenetic analysis studies and plant identification.
Figure 7. A sliding window analysis of nucleotide variability (Pi) values of different regions of
Toddalia.
X-axis: position of the midpoint of a window,
Y-axis: nucleotide diversity of each window.
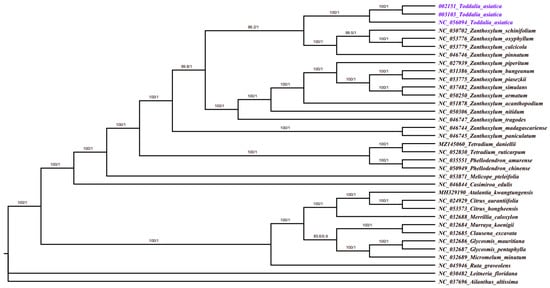
8. Phylogenetic Analysis
Plastomes are vital features for comprehending intra-and interspecific evolutionary histories, and recent studies have shown their potential in phylogenetic, evolution, and molecular systematic analysis
[18][30]. The more variable the sites in chloroplast plastomes are, the more potential they have in solving phylogenetic relationships within and among species
[41][42][53,54]. Phylogenetic study of the two data sets using 35 complete plastomes and 79 protein-coding genes formed a well-resolved phylogenetic tree. The maximum likelihood (ML) phylogenetic tree using complete plastomes was congruent with the Bayesian inference (BI) tree using protein-coding genes with a high support value in almost every branch (
Figure 8). The two samples (
T. asiatica 002152 and
T. asiatica 003103) collected from Kenya, Africa were a sister to
T. asiatica collected from China, Asia. These
Toddalia species formed a sister clade to
Z. calcicola,
Z. oxyphyllum,
Z. pinnatum, and
Z. schinifolium from the Southwest Pacific and East Asian species of
Zanthoxylum with a high support value. These results were in agreement with previous studies
[4][6]. The
Toddalia species analyzed in this study clustered together in the same clade and formed a sister clade with
Zanthoxylum species, thus showing a closer relationship of these species. Our results support previous studies
[4][2][6,8] of merging the genus
Toddalia with
Zanthoxylum. However, more cp plastomes of the
T. asiatica species are needed to understand the precise position of these species in the phylogeny. Inconsistencies in the topology of the tree may be attributed to low taxon sampling
[43][55].
Figure 8. Phylogenetic tree construction of 35 taxa using maximum likelihood (ML) and Bayesian inference (BI) methods using 79 protein-coding genes.
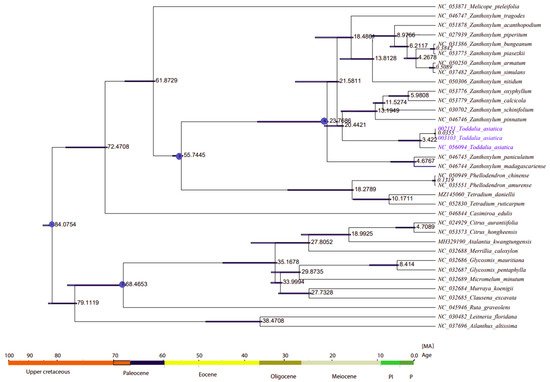
9. Divergence Time Estimation
The divergence time of Rutaceae was estimated at 85 million years ago (Mya) (HPD%)
[44][56] (
Figure 9, node 1). The two samples of
Toddalia species (
T. asiatica 002151 and
T. asiatica 003103) from Africa diverged from
T. asiatica from Asia 3.422 million years ago. These
Toddalia species diverged from the Southwest Pacific and East Asian species of
Zanthoxylum (
Z. pinnatum, (
Z. schinifolium,
Z. oxyphyllum, and
Z. calcicola)) at 20.4421 million years ago. Compared to the crown ages of the sister clade to
Toddalia,
Toddalia species are relatively recent and inadequate sampling of these species might have caused this difference. These
Toddalia species and Southwest Pacific and East Asian species of
Zanthoxylum formed a clade that diverged from the rest of
Zanthoxylum at 21.5811 million years ago while
Zanthoxylum species from Madagascar (
Z. paniculatum and
Z. madagascariense) diverged from the rest of
Zanthoxylum at 23.7608 million years ago. The genera
Zanthoxylum,
Toddalia,
Phellodendron, and
Tetradium are known as the oldest fossils of the family Rutaceae
[3][2][3,8]. The clade consisting of the genera
Phellodendron and
Tetradium diverged from
Zanthoxylum at 55.7445 million years ago. On the other hand,
Phellodendron diverged from
Tetradium at 18.2789 million years ago. Although these results shed light on the biogeography of
T. asiatica species, more of these species collected from a wide geographical area are needed in future studies.
Figure 9. Phylogenetic chronogram showing the evolutionary dating time taxa of the 15 genera in the family Rutaceae and two outgroups. The tree was estimated using Bayesian analysis of 79 protein-coding genes and 35 taxa in the Markov Chain Monte Carlo (MCMC) tree.